This article illustrates the potential of coupling a business process management system (BPMS) and LIMS, as presented in Part 1 of this paper. Part 2 highlights the distribution of tasks between BPMS and LIMS.
LIMS fulfills four requirements for BPM-based end-to-end workflow automation:
- The LIMS acts as a generic platform for recording the process data
- The LIMS provides extensive interaction components for human tasks
- The LIMS supports automated access to distributed process data within the automation system by specific services, if data acquisition is not handled by service tasks in the business process model and notation (BPMN) process
- The LIMS provides an integration platform. The integration refers to additional LIMS functions (enterprise application integration [EAI]) and interfaces to other information systems on the workflow level, e.g., master data management, document management, chemical inventory management, or data processing.
Coupling workflow control and documentation platform for automated biological
The focused BPM approach seeks to combine the functional synergy of LIMS and BPMS in a generic overall solution that enables the IT support of LIMS as required, synchronized with the execution of processes. In this context, IT support refers to selected activity-related software components. A representative selection of such support options and user interfaces for BPM applications in life science automation includes:
- Support of automated data processing
- Activity-related hierarchical process documentation, in the example: the experiment documentation with the plate layout used
- Recording variants for paper documents combined with a text editor
- Call for preconfigured process visualization of experimental results
- Activity-accompanying live visualization (subset of service area, i.e., virtual laboratory)
- Typical editor for chemical structures integrated in the master data management
- Optional activity support using voice recording or voice information, in particular, for cleanroom environments
- Activity-related user interface of a process documentation.
The BPMS initiates the documentation of the workflow within LIMS via service calls during the process execution. The services cover the communication and the data transformation. LIMS or BPMS supports human tasks with Web forms. Further service calls assume the horizontal systems integration of information systems on the workflow level, if necessary. They can also be directed to the vertically structured laboratory automation. Important tasks in application projects for the life sciences include automated pre- and postprocessing, such as test calculations, raw data processing, and statistical model calculations of experimental behavior. This functionality is also integrated in openLIMS (developed at the Center for Life Science Automation [Celisca] at the University of Rostock, Germany), and can be used via Web service interfaces. The included third-party systems are commercial statistical tools or common spreadsheets. The application described here is an example of other substance screenings regarding further targets that have been established at Celisca in order for the results to be generalized.
BPMN-based workflow automation—high-throughput screening of compound libraries
The biological screening of compound libraries is realized as a fully automated subprocess on a robot-assisted automation island. This platform is controlled by PCS SAMI® (Beckman Coulter, Brea, CA) (Figure 1). In addition to the laboratory robot (ORCA [Optimized Robot for Chemical Analysis], Beckman Coulter) used for sample and labware transport, the robot line integrates a Beckman Coulter Biomek 2000 liquid handler and several peripheral devices (shakers, incubators, bar-code print and apply, and photon counters [from several vendors]).
An additional in-house-developed software component expands the manufacturer’s platform via potential external access by way of Web service, and an asynchronous information provision via message queuing (e.g., for status query). Manual auxiliary activities for documentation, experiment preparation (providing the selected compounds and fitting the automation island with necessary labware), and experiment evaluation and interpretation complete the core automated process. Corresponding PCS methods and SOPs are available for the actual experimental processing of the biological assays.
 Figure 1 – Left to right: automation island for screening, mobile BPMS client, compound supply according to planning, fitting of robot line, and PCS method for control of the fully automated subprocess.
Figure 1 – Left to right: automation island for screening, mobile BPMS client, compound supply according to planning, fitting of robot line, and PCS method for control of the fully automated subprocess.With the device configuration described, one experimental run screened 80 compounds on four 96-well plates. For the screening of the whole compound library, multiple repetitions of the process in Figure 2 were necessary. Depending on the installation, throughputs of up to 1152 data points per day (in three runs) are possible.
Figure 2 shows the complete screening process as an executable BPMN process model. The business process consists of a set of activities that are performed together in an organizational environment. Distribution and coordination of work among people or systems is an important task of the BPMS. Thus, a BPMN diagram can be divided into pools and lanes to highlight the organizational structures involved (i.e., departments, employees, complex devices, and automation systems). The user tasks are assigned according to the required competencies to the roles “biologist” (upper lane in Figure 2) and “lab assistant” (lower lane in Figure 2) by placing them in the respective lane. Sequence flows that cross lanes indicate handover of work.
 Figure 2 – Executable process model of subprocess, HTS screening with GSK-3ß assay.
Figure 2 – Executable process model of subprocess, HTS screening with GSK-3ß assay.Automated planning
The biologist initiates the process by planning a new experiment within the LIMS, but without direct interactive access to the LIMS. The authentication information and experiment-related data are requested via Web forms: LIMS template to use for documentation, experiment name, number of test plates, and placement in the documentation hierarchy (project, study, etc.). For this, the required data are queried from the LIMS by appropriate Web services and are provided to the user in drop-down lists. Based on these data, documentation of the current experiment is created by utilizing a further function of the openLIMS Web service based on the selected template. This Web service returns a list of identification data (ID) that are used as bar codes for the test plates.
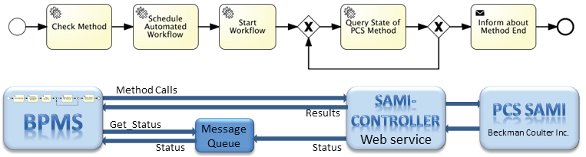 Figure 3 – Communication scheme between BPMS and PCS-controlled lab robot system (bottom) and corresponding BPMN subprocess for integration of the PCS (top).
Figure 3 – Communication scheme between BPMS and PCS-controlled lab robot system (bottom) and corresponding BPMN subprocess for integration of the PCS (top).The use of unique ID for the experimental run and test plates is essential for the subsequent automated assignment of raw data to the corresponding experiment documentation into the LIMS. Concurrent to the bar-code labeling performed by the laboratory assistant, the biologist can continue the detailed experiment planning with the selection of test compounds. For this, the corresponding web page of openLIMS is provided. The automated provision of the appropriate modules saves time and manual effort. The steps previously required to navigate within the project hierarchy of the LIMS to the documentation of the current experiment are no longer needed. This can avoid allocation errors.
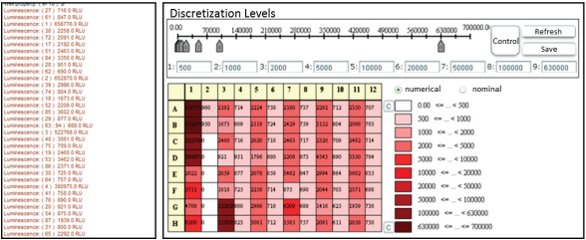 Figure 4 – The
Figure 4 – The open
LIMS provides several visualization methods (e.g., heatmap of a single test plate, right) that are common practice in life science applications, which allows a quick assessment of the results (left).Experiment preparation
Time-critical human tasks, such as the provision of compounds, are logged in the LIMS with a date-time stamp by the workflow control immediately after the confirmation statement is given by lab personnel. The confirmation statement is an important element of quality assurance in the case of manual tasks. It allows one to determine how long the compounds were stored in less than optimal conditions until they were processed. Further parameters (date-time stamp) are added to the experiment documentation using a function of the LIMS Web service. If compounds were provided in sufficient quantity in accordance with the plan, an automated log of their withdrawal from inventory is generated. Interactive intervention by the biologists is only necessary when changes are necessary due to missing stocks.
Automated screening
The fully automated subprocess—Screening—includes sample preparation and the actual execution of the screening. The laboratory robot system is controlled by the PCS SAMI platform with corresponding proprietary method. To perform this method by the PCS, several steps are necessary: check availability of the method, schedule with current parameters, and start the method. These steps are encapsulated in the corresponding BPMN subprocess model as service tasks with an implemented Web service (Figure 3). In cases of status changes, the SAMI controller middleware writes the current status in a message queue. This asynchronous communication significantly reduces the communication load on the PCS. The values “done/error/canceled” signal the final states of the subprocess that determine further process flow. It is possible to monitor the execution via Web cam. The persons involved receive a link to the live stream via a work list.
Automated postprocessing
The normalization of raw data is an event-triggered data processing service task. The open data processing integrated in openLIMS is based on a common spreadsheet tool (e.g., MS Excel, Microsoft Corp., Redmond, CA). It requires corresponding processing macros and a prepared template with the calculation formulas and algorithms. The processing macros define the data transfers to and from the template sheet. The data processing module is provided by openLIMS via Web service as with other functionalities. Automation of these processing steps relieves the scientist of monotonous activities that need to be continually performed (in the case of the above example, they are performed separately for each of the four test plates) and helps to avoid errors. The automatically aggregated data allow evaluation of biological activity of the tested compounds.
The documentation of the experiment run within the LIMS links it with the raw data, the normalized results, as well as the characteristics of the tested compounds, including the documented manufacturing information and storage conditions. All data that are relevant for archiving and interpretation of results are recorded consistently and are context sensitive. They are transparent and available at any time for research.
Evaluation and further processing of data
An e-mail sent by the workflow automation to the biologist responsible informs about the results of the current experiment and links a visualization of the results using the embedded functionality of openLIMS (Figure 4). This aggregated information is generated with direct database queries, and makes it possible to assess the experiment results. It allows the prioritization of the next evaluation steps in the work list of the biologist.
Effects of BPM-driven workflow automation
The complex application example presented points out the typical interplay between the BPMS-based workflow automation and a domain-specific information system. In addition to the higher level of automation, it results in workflow-driven documentation of process data. Personnel will be relieved of documentation tasks, data transfers, and transformations.
With regard to the rules of Good Laboratory Practice (GLP), in addition to systematic research and development, the BPM approach improves quality management. The close integration of recording information system and workflow automation leads to less redundant data storage, because the resulting data will be provided to the target system immediately and in the required form. Alternatively, the data are stored uniformly in the central information system.
The benefits of BPM technologies are particularly apparent when complex and changing end-to-end processes involve control, recording, and monitoring. The process modeling using the graphical notation standard BPMN 2.0 meets the requirements of high process transparency for both robust workflow controls and knowledge management. The process transparency in life science laboratories extends to any detailed processes of the hierarchical automation system in the lab and all necessary professional networking of the interdisciplinary teams involved. Inclusion of all human tasks in the controlled workflow increases the robustness and quality of the processes. The control and dedicated just-in-time IT support for manual subprocesses benefit the BPM-assisted workflow automation independent of the degree of automation of the laboratory systems used.
References
- OMG: Business Process Model and Notation (BPMN) Version 2.0; www. omg.org/spec/BPMN/ 2.0; 2011.
- Allweyer, T. BPMN 2.0. Introduction to the Standard for Business Process Modeling; Books on Demand: Norderstedt, 2010.
- Silver, B. BPMN Method and Style; Cody-Cassidy Press: Aptos, CA, 2009.
- Clark, W. LIMS product round-up. A look at the latest innovations in LIMS. Laboratory Informatics Guide 2012, 10–14.
- Thurow, K.; Göde, B. et al. Laboratory information management systems for life science applications. Organic Process Research & Development: an international journal published jointly by the American Chemical Society and the Royal Society of Chemistry, 2004, 8, 970–82.
- Göde, B.; Holzmüller-Laue, S. et al. Flexible IT-plattform zur automatisierten HTS-wirkstoffanalyse. GIT Laborfachzeitschrift 2007, 5, 741–4.
- Holzmüller-Laue, S. et al. A highly scalable information system as extendable framework solution for medical R&D projects. In: Adlassnig, K.-P., Ed.; Medical Informatics in a United and Healthy Europe. Proceedings of MIE 2009, XXIInd International Congress of the European Federation for Medical Informatics; IOS Press: Amsterdam, 2009, 101–5.
Silke Holzmüller-Laue, Ph.D., is Senior Researcher, and Kerstin Thurow, Ph.D., is CEO, Center for Life Science Automation at the University of Rostock, F.-Barnewitz-Str. 8, 18119 Rostock, Germany; tel.: +49 381 4987721; fax: +49 381 4987702; e-mail: [email protected]. Bernd Göde, Ph.D., is Group Leader, Institute of Automation, University of Rostock, Germany.