Drug development today is
a time-consuming and
expensive business, with
costs reaching over $1 billion
from drug discovery to market.1
The cost escalates during clinical trials;
therefore, it is of the utmost importance
to avoid late-failing drugs by
selecting the best candidate early in
the process. Traditional biosensors are
important and powerful tools in the
drug discovery process for their ability
to measure the pure interaction
between a drug candidate and its target.
However, most biosensors are limited
to using a purified target molecule
immobilized to the sensor surface. This
oversimplifies the biological context
and presents an incomplete picture of
the in vivo situation.
Eukaryotic membrane proteins such
as G protein-coupled receptors
(GPCRs) and ion channels are the
preferred target for more than 60% of
current therapeutic drugs.2 These biomolecules
need a lipid bilayer to maintain
their structure and function and
therefore cannot be easily isolated and
studied with traditional biosensor methods.
Conventional cell-based assays, on
the other hand, are well suited for studying
the effect interactions have on cells,
but do not disclose the full dynamics of
the interaction itself.

Figure 1 - Attana Cell 200 biosensor.
The Attana Cell™ 200 biosensor
(Attana AB, Stockholm, Sweden) measures
label-free, full kinetics in real time
with the target in its biological context
(Figure 1). Cells can be grown directly
on the sensor surface, and the interacting
biomolecule is introduced to the
continuous flow system as in a traditional
biosensor. The instrument is also compatible
with standard biosensor surfaces
and can therefore be used to compare the
binding to a purified target with that of cellular interactions. The combination
of the QCM technology and cell sensor
chip results in biologically relevant biosensor
measurements.
QCM technology
QCM (Quartz Crystal Microbalance)
technology enables studies of molecular
interactions by measuring the weight of
the molecules, much like a very sensitive
scale or balance. When molecules are
added to or removed from the sensor surface,
it is detected as a change in the oscillation
frequency of the sensor crystal; the
change in resonance frequency is correlated
to the change in mass on the surface.
QCM technology does not have the same
limitations with regard to surface proximity
as other biosensor technologies, making
it possible for the instrument to measure
binding to large structures such as cells.

Figure 2 - Cell sensor chip.
Cell sensor chip
The cell sensor chip (Figure 2) permits
the user to culture mammalian cells
directly on the cell-optimized polystyrene
sensor surface. A cell suspension is added
to the cultivation chimney and the cells
are allowed to adhere to the surface. The
growth and cell density can be validated
using fluorescent microscopy. By replacing
the cultivation lid with a measurement
lid, the sensor chip can be docked
in the instrument and a biosensor experiment
measuring interactions with targets
on the cell surface can be started.
Molecular interactions in a
biologically relevant context
The cell-based biosensor can be used to study
membrane proteins in their natural environment,
the cell membrane. This can be
achieved in several ways, from immobilizing
membrane preparations on the sensor surface
to studying interactions directly on cells. Two examples are presented that cover the range of application possibilities,
the first working with lipoparticles and the second with cells.
Molecular interactions with lipoparticles
Lipoparticles composed of natural cell membrane are both
durable and stable and as such are well suited for studying
molecular interactions. In this application example, 150-nm-diam
lipoparticles (Integral Molecular, Philadelphia, PA)
incorporating the GPCR chemokine receptor 4 (CXCR4)
were used to study the interaction between CXCR4 and an
anti-CXCR4 antibody. The particles containing CXCR4 were
immobilized onto Attana Biotin Sensor Surfaces (Attana AB)
using memLAYER reagents (Layerlab, Gothenburg, Sweden).
The reagents are based on the company’s proprietary Tethered
Enhanced Liposome Immobilisation (TELI) technology and
consist of a cholesterol–DNA strand that is naturally integrated
in the lipid bilayer and a complementary biotin–DNA
strand that is immobilized onto a NeutrAvidin-coated sensor
chip (Pierce, Rockford, IL). DNA hybridization then enables
stable capture of lipoparticles on the sensor surface.
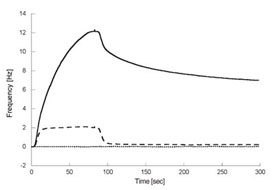
Figure 3 - Anti-CXCR4 antibody binding to lipoparticles using the cell-based
biosensor. Comparison of anti-CXCR4 antibody binding to three different surfaces:
lipoparticles containing CXCR4 (solid line), lipoparticles without CXCR4
(dashed line), and a control surface with immobilized biotinylated bovine serum
albumin (BSA, dotted line). The antibody is injected for 84 sec over the three
different surfaces and the dissociation of the antibody is monitored for 200 sec.
The antibody displays high-affinity
binding to lipoparticles containing CXCR4,
whereas weak and off-target binding to lipoparticles without CXCR4 was
detected. No binding to the control surface was detected.
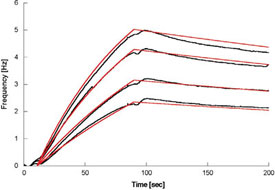
Figure 4 - CXCR4 antibody binding to lipoparticles fits a 1:1 binding model.
Referenced binding data, obtained by subtracting the binding response to lipoparticles
without CXCR4 from that of lipoparticles containing CXCR4 (black), are
fitted to a 1:1 binding model (red).
The interactions between CXCR4-containing lipoparticles
and anti-CXCR4 antibody were studied in real time using the
cell-based biosensor. The results show both specific interaction
and off-target binding to the cell membrane or membrane proteins
(Figure 3). No binding was detected to biotinylated BSA
directly immobilized onto a control surface. The antibody interactions
with CXCR4-containing
lipoparticles display a dose
dependency. In addition, by using data from antibody interaction
with lipoparticles lacking CXCR4 as a reference, an investigation
of only the specific interactions is possible (Figure 4). A
1:1 binding model was used to fit (red lines) the experimental
data (black lines), and binding rate constants calculated. The
affinity of the interaction was determined to be 2.2 nM.
Molecular interactions with cells
Combining the high specificity of monoclonal antibodies to
their targets with the pharmacological potency of cytotoxic
drugs can generate a higher therapeutic efficacy and is often
used in oncology.3 There are currently more than 15 promising antibody–drug conjugates (ADC)
in clinical trials, and there is also an
increased focus in early drug development
with many promising candidates.4
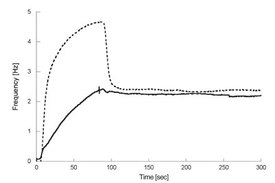
Figure 5 - Comparison of the binding of Herceptin (solid line)
and modified Herceptin (dotted line) to cells expressing HER2
using the cell-based biosensor. Herceptin is injected for 84 sec and
the dissociation of the antibody is monitored for 200 sec. Modified
Herceptin displays both specific off-target and/or nonspecific binding
to cell surface components, which is indicated by the two concurrent
types of reactions, one high-affinity, specific interaction with relatively
slow on and off rates (also present when injecting unmodified
Herceptin) and a second low-affinity, off-target interaction with
fast on and off rates.
In an attempt to selectively trigger cancer
cell death with an ADC, an Attana
biosensor user modified the anti-HER2
breast cancer marker antibody Herceptin
(Genentech, South San Francisco, CA),
and needed to verify that the binding
characteristics were unchanged.5 In a
traditional biosensor experiment with
the purified target immobilized on a
sensor surface, the modified Herceptin
kinetics were unaffected. However,
when measuring binding to the target in
its natural environment, the cell membrane,
the results were quite different
(Figure 5). The modified Herceptin had
retained its specific binding to the target,
but off-target binding to the membrane
or other components in the membrane
was evident. The off-target interaction
is fast but is vital for understanding the
function of the modified antibody in the
biological context.
Summary
Keeping costs down and speeding up
the drug development process is an
important challenge for
drug companies. One central
component is to select
the best drug candidates
during each step of the
development process. The
information gained from
measurements with the
Attana Cell 200 biosensor
can be used to select better
candidates earlier, therefore
saving both time and money
in consecutive preclinical
and clinical trials.
The biosensor delivers more
biologically relevant information
by providing the possibility
of measuring molecular
interactions with cells in real
time and without the use of
labels. It also shows the full
dynamics of the interaction,
including off-target interactions
with other components
on the cell surface and nonspecific binding,
which are important for understanding
the in vivo process.
References
-
Landers, P. Cost of Developing a New
Drug Increases to About $1.7 Billion. Wall
Street Journal, Dec 2003.
- Overington, J.; Al-Lazikani, B. et al. How
many drug targets are there? Nature Rev.
Drug Discov. 2006, 5, 993–6.
- Kovtun, Y.; Goldmacher, V. Cell killing
by antibody–drug conjugates. Cancer Lett.
2007, 255, 232–40.
- Beck, A. The next generation of antibody–drug conjugates comes of age. Discovery
Med. 2010, 10(53), 329–39.
- Kovacs, A. Molecular interaction studies
in cells: exploring biology at a new level.
Innovations Pharm. Technol. Jun 2010,
42–6.
Ms. Elovsson is Product Manager, and Dr.
Aastrup
is CEO, Attana AB, Björnnäsvägen 21,
114 19 Stockholm, Sweden; tel.: +46 8 674 57
12; e-mail: [email protected]. Dr. Pei
is a Professor, College of Science, Northwest A&F
University, Yangling, People’s Republic of China.