Glycoxidative modification of proteins involving free radical intermediates and reactive carbohydrates leads to complications in diabetes, atherosclerosis, and Alzheimer’s disease.1 Similar modifications of the common nucleobases are also implicated as a major step in the occurrence of cancer. Modification of proteins involves reactive amino acids such as lysine, arginine, and histidine residues.
Investigations of protein modifications have led to the discovery of important biomarkers such as carboxymethyllysine (CML),2 carboxyethyllysine (CEL),3 and pentosidine4 compounds. These biomarkers are used extensively to assess the damage to proteins due to diabetes and other age-related diseases. Lysine (Lys) and arginine (Arg) amino acids are involved in protein glycoxidation and lipoperoxidation reactions; therefore any other biomolecules having NH2 and =NH groups like those in Lys and Arg may also be susceptible to similar glycoxidative and lipoperoxidative reactions. Since DNA bases contain NH2 and =NH groups, nucleotides containing such groups present potent centers of modification by glycoxidative and lipoperoxidative free radical reactions.5,6
Some carcinogens react with DNA at several different sites, and the formation of more than one product is possible.7 This is supported by Ref. 8, which reports the occurrence of N2-carboxymethyl-2’-deoxyguanosine (CMdG) in calf thymus DNA and cultured human kidney epithelial cells exposed to induced glyoxal modification. Those authors indicated that the identified CMdG derivative was stable at physiological conditions of temperature and pH over long periods. Carboxymethyl-2’-deoxycytidine (CMC) was detected from in vitro reactions of DNA precursors with diazoacetate.8 The authors claim discovery of carboxymethyl-2’-deoxyadenosine (CMA) and CMC as the first report of this kind,9 and is disputed here by reported detection of these compounds in Refs. 5 and 6.
In the HPLC analysis performed in this study, an HPLC system from Shimadzu Scientific Instruments (Columbia, MD) was used. The HPLC system consisted of an LC-20AT low-pressure gradient pump with DGU-20A5 on-line degasser, SIL-20AC autosampler, HYPERCARB 5 μm 150 × 4.6 mm reversed-phase column, HYPERCARB 5 μm 10 × 4 mm guard column attached to the column head, CTO-20A column oven, and CBM-20Alite system controller. An SPD-20AV UV-VIS detector at a wavelength of 254 nm was used.
Mitochondria and nuclei, which are generally recognized as the two major targets of oxidative stress, also contain a variety of DNA repair enzymes that take appropriate measures to correct the oxidant-induced DNA modification.10,11 Damage most likely occurs when the endogenous antioxidant network and DNA repair systems are overwhelmed.12–14 However, it is essential for the cells to repair DNA damage in order for them to function normally. Refs. 15 and 16 cite experiments in which 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-OxoG) was identified as a marker of reactive oxygen species (ROS) induced DNA damage. The experiments indicated an increase in 8-OxoG levels in the tissue of diabetic rats and in the urine of patients with type 1 and type 2 diabetes.16–18
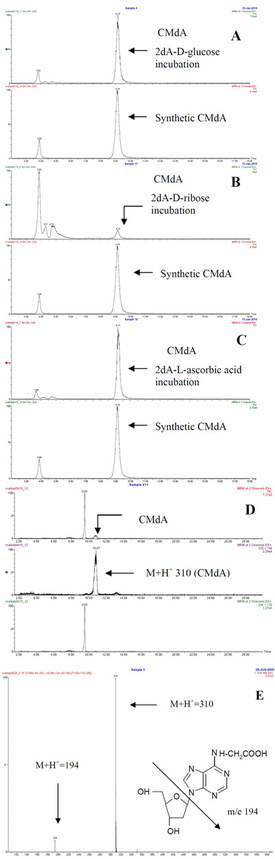
Figure
1 - Electrospray LC-MS spectrum of dA with a) D-glucose, b) D-ribose,
c) L-ascorbic acid, d) urine sample, and e) total ion chromatogram (TIC)
and mass spectrum of CMdA from synthesis. Electrospray LC-MS spectrum
of dA with D-glucose, D-ribose, and L-ascorbic acid incubated samples
gave a CMdA peak at RT 9.1 min, a molecular ion peak at M+H+ 310, and another major peak at m/e 194. The CMdA peak at RT 9.1 min matched well with the synthetic spectrum analyzed.
Detection and quantification of DNA adducts are very important in relation to diseases such as cancer and possibly diabetes. Both high sensitivity and high selectivity are required to detect DNA adducts, because the content of adducts in DNA may be very low compared with those of normal bases, and only small amounts of DNA are typically available for analysis.19 An investigation of the mechanisms expressing toxicity for each DNA adduct is needed in order to
cure or at least mitigate some of the age-activated diseases and also to determine the toxicity.19 Diabetes-associated modifications of DNA by ROS may contribute to diabetic complications and to the generation of cancer cells by mutations of the genes present in the DNA strand.
Experimental
Chemicals and samples
All chemicals were obtained from Fisher Scientific (Pittsburgh, PA) unless otherwise indicated. Human serum DNA and fasting urine samples were collected from male and female subjects.
Methods
1. Incubations of 2’-deoxyadenosine (dA) with D-glucose, D-ribose, and L-ascorbic acid: A mixture of dA (13 mg, 0.05 mmol) and D-glucose (9 mg, 0.05 mmol) was incubated in 0.2 M phosphate buffer (1 mL, pH 7.4) (two drops of toluene were added to prevent fungal growth) at 37 °C for 30 days. The reaction mixture was kept constant at pH 7.4 during the incubation period by adjusting the pH as required; 100-μL samples were collected at different time points (2, 4, 6, 8, 10, 20, and 30 days) during the incubation period and stored in the refrigerator before analysis. For analysis, a typical sample was adjusted to pH 7.0 and dried (<60 °C) in a rotary evaporator. Methanol (3 mL) was added to the dried residue; the mixture was sonicated, vortexed, and centrifuged; and the methanol fraction was collected. The sample was then subjected to HPLC and electrospray LC-MS. Figure 1a–e show a molecular ion (M+H+ 310) and fragment m/e of 194. Incubations of 2’-deoxyadenosine (13 mg, 0.05 mmol) with D-ribose (7.5 mg, 0.05 mmol) and L-ascorbic acid (8.8 mg, 0.05 mmol) were carried out in separate experiments in 0.2 M Pi (1 mL), pH 7.4, at 37 °C for 30 days, and methanol extracts were subjected to HPLC and LC-MS. Figure 1b and c show a molecular ion (M+H+ 310) and fragment m/e of 194.
2. Synthesis of CMdA using dA and iodoacetic acid: Incubation of 2’-deoxyadenosine (13 mg, 0.05 mmol) and iodoacetic acid (9 mg, 0.05 mmol) in 0.2 M Pi (1 mL), pH 7.4, 37 °C for 7 days was also carried out. Aliquots (200 μL) were collected, and the methanol extracts from pH 7.0 adjusted samples were analyzed by HPLC and LC-MS. Figure 1e shows a molecular ion (M+H+ 310) and fragment m/e of 194 as in experiment (1) above.