In the past decade we have seen advances in laboratory
automation and in assays used to assess biological endpoints. At the Southern Research Institute
(Birmingham, AL), these new technologies have
greatly enhanced its high-throughput screening (HTS) capabilities for viral targets in cell-, protein-,
and enzyme-based assays. These technologies can be
leveraged to rapidly screen large compound libraries
to discover new antivirals for previously unrecognized,
emerging pathogens. There has been a dramatic
increase in emerging pathogens, which impact
public health and can result in serious economic
impact at regional or global levels.1,2 Emerging viral
illnesses present a number of unique challenges in
drug discovery efforts, including limited reagents,
access to viral strains, containment, and availability
of trained scientists. In the last decade, resources
have become available for the development of facilities
to study these pathogens, and importantly, for
the development of therapeutics for the treatment
of the diseases they inflict. This article presents the
special issues that must be considered in the creation
of a drug discovery program for these types of
viral pathogens, with examples from the Southern
Research Institute drug discovery research program
in Biosafety Level 2 (BSL2) and Biosafety Level 3
(BSL3) containment.
Special considerations for pathogens
The National Institutes of Health (NIH, Bethesda,
MD) and Centers for Disease Control (CDC,
Atlanta, GA) have three categories for emerging
and biodefense-related pathogens: A, B, and
C.3,4 These pathogens are grouped by the ease
with which they can be spread and the severity
of the disease they cause. For example, many of
the hemorrhagic fever diseases, such as the Ebola
and Machupo viruses, and the bacterium Bacillus
anthracis (anthrax), are placed in Category
A, because they are thought to have the highest
risk and impact in spread and illness. Nipah
virus, influenza, severe acute respiratory distress
syndrome coronavirus (SARS CoV), and hantaviruses
are grouped as Category C agents, and are
considered emerging infectious disease threats.
In addition to the basic research efforts needed
to understand the life cycles of these pathogens
and their effect on human or wildlife hosts, it is
essential that we begin to develop therapeutics for
the treatment of the diseases they inflict. The vast
majority of these agents require specialized containment
laboratories such as BSL3 or 4.5
Several important matters must be considered when
establishing a facility dedicated to the discovery and
development of new therapeutics, i.e., the requirement
for BSL3 or 4 laboratory facilities, regulatory
issues (e.g., Select Agent Rule,6 GLP compliance,
and CDC and USDA permits), personal protective
equipment (PPE), and containment of equipment
as necessary. In addition, the processes for flow of
work among the various laboratories must be thoroughly
reviewed to ensure high-quality results. The Southern Research Institute’s overall approach to
the implementation of HTS for BSL2 and 3 viral
pathogens7,8 is outlined below.
Phase 1: Assay design and development

Figure 1 - Steps undertaken through hit validation. The four phases, with various steps, from assay design and development through
hit validation.
The first HTS assay implemented at Southern
Research was for the SARS CoV.9 The luminescent-based assay measures the inhibition of SARS
CoV-induced cytopathic effect (CPE) in Vero E6
cells. The following section describes the various
steps undertaken through hit validation (Figure
1). The first step in the process was a risk assessment
of the pathogen with respect to its transmissibility
and the type of containment required
within the BSL3. Hence, prior to the development
of the screen, decisions were made on the
type of PPE to be worn by laboratory workers
(Figure 2) and type of containment required
for equipment that might aerosolize virus.
The coronaviruses (order Nidovirales—family
Coronaviridae—genus Coronavirus) are members
of a family of positive-sense RNA viruses that
replicate in the cytoplasm of animal host cells.
They are a diverse group of large, enveloped
viruses that infect many mammalian and avian
species, causing upper respiratory, gastrointestinal,
hepatic, and central nervous system diseases.
It is generally accepted that SARS CoV
is transmitted by close person-to-person contact
through coughing or sneezing, which deposits
respiratory droplets on the mucous membranes
of the mouth, nose, or eyes of persons nearby.
Additionally, transmission can occur through contact with surfaces contaminated with the infectious
droplets.
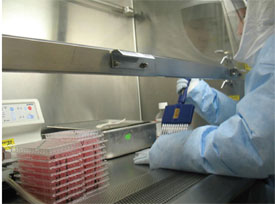
Figure 2 - A scientist at Southern Research conducts in vitro efficacy
testing of a vaccine candidate to prevent high path Avian influenza
(HPAI) infections. This work is being conducted in the company’s BSL3
laboratory in Birmingham, using both hand screening and robotic screening
measures.
Because SARS CoV is airborne and has the ability to
be transmitted from person to person, it was decided
that those handling the virus would wear positive
air-pressure respirators (PAPRs), full-length Tyvek®
suits (DuPont Co., Wilmington, DE) with a wraparound
gown over the scrubs, double shoe covers, and
double latex gloves (Figure 2). Workers dressed in
the wraparound gown and additional layer of gloves
and shoe covers before entering the HTS laboratory
within the BSL3, and removed them when exiting the HTS laboratory. The liquid handling equipment
that dispenses the virus into drugged plates
was placed in a biosafety cabinet (BSC). However,
the plate reader for reading the endpoints was kept
outside. The plate reader and its plate stacker will
not fit in a BSC, and while there is inherent risk of
plates dropping or becoming jammed in the reader, it
was decided that the PPE and BSL3 would act as sufficient
barriers to any possible accidental exposure.
Once the protective barriers were in place, the next
step in the establishment of the screen was to define
the optimal flow of work (Figure 3). While it would
be advantageous to add chemical compounds to the
screen after virus addition, the problems associated
with maintaining compound libraries in the BSL3
would be formidable. In addition, because of BSL3
regulations, compounds cannot leave the facility
without undergoing a decontamination procedure,
which could alter the integrity of the sample. It was
agreed that storage and aliquoting of compounds
would take place 24 hr after the addition of cells
to multiple-well plates. All of the processes, procedures,
and safety considerations were submitted to
the Institutional Biosafety Committee for review.
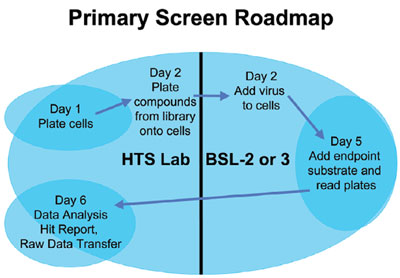
Figure 3 - Optimal flow of work.
For the BSL2 virus HTS such as for influenza, the
majority of the safety guidelines above continue to be
followed. The major difference lies solely in the PPE.
In the BSL2 laboratory, gloves, laboratory coats, and
safety goggles are worn. All of the equipment safety
barriers are the same as described for BSL3.
Once approval was met, the actual development and
optimization of the assay and evaluation of the flow
of work outlined was undertaken. Optimization of
the assay requires judicious attention to every detail,
such as the choice of consumables, cell age, growth
conditions, and liquid handler. For example, using an
eight-channel peristaltic pump, Vero E6 cells were
plated in black, clear-bottom, 96-well plates at a
density of 10,000 cells/well in a 50-μL assay medium.
Careful attention needs to be paid to controlling the
humidity of the CO2 incubator, and some brands
appear to do this better than others. The next day,
25 μL of compounds were added to the cells using a
flexible liquid handling system in a biosafety cabinet.
In this case, the BSC was used to protect the cells
from the environment, i.e., keep them in an aseptic
environment. In the Southern Research work flow,
plates are drugged and then hand-carried into the
BSL3 in groups of 50 plates per run. Using more than
50 plates is difficult in the BSL3 environment simply
because of movement logistics. In the BSL2 HTS
virology laboratory, however, up to 125 plates can be
moved effectively, since the constraints of BSL3 are
not present. Plating of the compounds requires about
twice as much time as plating of the virus; thus the flow of work depends on the equipment chosen
for this activity. In all instances, compounds
are plated prior to virus addition.
After incubation for three days, a luminescent
cell viability reagent is added that
measures cellular adenosine triphosphate
(ATP) levels to each well using a laboratory
automation workstation benchmark system.
Recently, the laboratory switched to a system
with a smaller footprint. Smaller footprints
in equipment are of particular advantage
in BSL3, where space is at a premium. For
96-well assay plates, plates are shaken on a
compact benchtop plate shaker, which has
a small footprint as well. For 384-well plates,
the plates are not rocked. Luminescence is
measured using a plate reader. An Ethernet
interface allows direct uploading of the data
into the database and analyses using an integrated
data management software application outside
of the BSL3. Each plate contains negative and
positive controls that are used to fail or pass plates
that have Z-values of less than 0.5.9
Phase 2: Assay validation
As part of the validation for the viral HTS assay,
small compound libraries are used to determine
the reproducibility of the hits and the hit rate. A
“hit” is identified as an active compound that either
activates or inhibits the assay signal above a defined
threshold value from the sample mean signal. A hit
for this assay was considered to be any compound
exhibiting a % CPE inhibition of >50%. The hit
rate can vary depending on the virus.9,10 While
the library in this study was not validated against
a high and low drug concentration, subsequent
studies with other viruses have shown it to be an
important step at this stage of assay validation. For
example, in the validation effort of the assay to discover
small molecules that inhibit the cytopathic
effect exerted by influenza viruses, the laboratory’s
high-throughput cell-based assay was used to screen
16,000 compounds from a commercially available
compound library at a low and high concentration.
10 The hit rate for the compounds screened at
low concentration was 0.02% compared to 0.38%
at the high concentration, for an approximately
19-fold increase in the hit rate. For the SARS CoV
screen, hit rates of 0.01%9 were observed.
A comparison of the screening results from the same
set of compounds assayed on different days is the
most effective way to determine the reproducibility
of the assay. If one relies solely on the control wells
to assess the assay, one may develop a false sense of
confidence in its capabilities. The liquid handling
steps used for the control wells, which are usually
on either end of the plate, often differ from the steps
used for the unknowns in the compound library.
Also, the positive control compounds are usually
limited to one or two mechanisms of action. Reliance
only on data from these compounds may inadvertently
bias one against compounds operating by
different mechanisms.
While two libraries comprising known biologically
active compounds were used for the initial validation,
another rationale for choosing the validation
library is that one can select a subset of compounds
representing the chemical diversity of the larger
compound library. By doing this, the hit rate at a
proposed screening concentration can be estimated,
allowing one to adjust the concentration up or down
for the large screen. A benefit to choosing the validation
set in this way is that when the large library
is screened, the validation compounds are scattered throughout the large screen. This provides an independent
third assay of these compounds that can be
used to assess the quality of the large run.
Another advantage of choosing a large compound
validation set relates to troubleshooting the automation.
Typically, running a few plates at a time is
insufficient to identify problems that are likely to
occur when handling a large number of plates in
one day. The optimized validation set of Southern
Research is 10,000 compounds, or about 10% of the
large library.
Phases 3 and 4: Compound library screen and hit validation
At this stage, the general algorithm for HTS antiviral
screening employs the validated primary cell-based
assay for determining single-dose efficacy with
one or more of the laboratory’s large libraries. This
type of approach has been used successfully to screen
nearly two million compounds. This is followed by a
second, similar cell-based screen for toxicity evaluation
of the compound, which showed 50% or better
activity and dose response (DR) confirmation.
Those compounds with robust SI values are confirmed
in secondary assays (such as a plaque assay).
As an example of the large compound library screen,
100,000 compounds were screened in duplicate from
the 100,000-compound library against SARS CoV.9
The hit rate for this library (compounds that inhibited
CPE by >50%) in a single-dose (SD) format was
determined to be approximately 0.8%.
To screen selected hits from the SD assay, compounds
were confirmed in a DR format. Every HTS
assay has a false positive rate, and hits from the
primary SD screen are only considered potential
positives until that activity is confirmed in a second
DR screen, as mentioned above. Hence, the SD
screen identifies a smaller subset of effective lead
compounds (i.e., hits), which are then run through
one or several secondary assays to define the compound
dose response and/or toxicity. The accuracy
of the primary assay is paramount for identifying hits,
while simultaneously reducing the number of compounds
funneled through the secondary assays and
greatly decreasing the amount of time and resources
required to evaluate lead antiviral compounds. The
criterion for determining compound activity is based
on its selective index (SI50). The compounds with
an SI50 value of <4 are classified as not active, SI50
= 4–9 as slightly active, SI50 = 10–49 as moderately
active, and SI50 >50 as highly active. Following confirmation,
structure–activity relationships are defined
using a cheminformatic approach to identify other
similar molecules that might be available commercially.
Additional parallel chemical synthesis may
be done to generate a group of structurally related
compounds, which are reevaluated with the aforementioned
DR assays and further characterized for
mechanism of action if promising.
Next, an assay is used to determine the point in the
SARS CoV life cycle that the DR hits inhibited. In
effect, this simple screen allows one to ascertain if
the inhibition of the compound was early (entry)
versus late (replication). The laboratory’s biochemical
assays also exploit the high sensitivity of fluorescence detection, or use proximity assay formats with
ultralow background interference.
The development of a panel of biochemical assays
is important for the identification of the mode of
action for each lead candidate. These assays target vital components of the virus life cycle that are necessary
for infection, disruption of host–cell mRNA
translation, transcription, and virus budding. The
targets are often unique to the virus, and similar
activities are not found in the host such as for influenza,
which has neuraminidase, M2 proton channel,
and NS1A RNA binding activities.
Summary
The activities described above give an introduction
to the first phase of activity essential to drug discovery,
and this is clearly just the beginning. Once a hit
has been verified using a combination of cell-based
and biochemical methods, we can move forward to
determine the toxicity and efficacy of the drug in an
animal model. It is after this stage that we will gain
valuable insight into the suitability of the compound
class for lead optimization medicinal chemistry and
follow-up preclinical toxicology testing. It is hoped
that recent advances in infrastructure, new assays,
and HTS procedures will lead to the discovery of
new antiviral drugs for emerging and reemerging
pathogens, those of which we are aware and those
that are yet to come.
References
- Satcher, D. Emerging infections: getting ahead of the curve. Emerg. Inf. Dis. 1995, 1, 1–6.
- King, D.A.; Peckham, C.; Waage, J.K.; Brownlie, J.; Woolhouse, M.E.J. Infectious diseases: preparing for the future. Science2006, 313, 1392–3.
- www.bt.cdc.gov/agent/agentlist-category.asp.
- www3.niaid.nih.gov/biodefense/bandc_priority.htm.
- www.cdc.gov/OD/OHS/biosfty/bmbl4/bmbl4toc.htm.
- www.cdc.gov/od/sap/.
- Peiris, J.S.; Lai, S.T.; Poon, L.L.; Guan, Y.; Yam, L.Y.; Lim, W.; Nicholls, J.; Yee, W.K.; Yan, W.W.; Cheung, M.T.; Cheng, V.C.; Chan, K.H.; Tsang, D.N.; Yung, R.W.; Ng, T.K.; Yuen, K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet2003, 361, 1319–25.
- Kuiken, T.; Fouchier, R.A.M.; Schutten, M.; Rimmelzwaan, G.F.; van Amerongen, G.; van Rie, D.; Laman, J.D.; de Jong, T.; van Doornum, G.; Lim, W.; Ee Ling, A.; Chan, P.K.S.; Tam, J.S.; Zambon, M.C.; Gopal, R.; Drosten, C.; van der Werf, S.; Escriou, N.; Manuguerra, J.-C.; Stöhr, K.; Malik Peiris, J.S.; Osterhaus, A.D.M.E. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet2003, 362, 263–70.
- Severson, W.E.; Shindo, N.; Sosa, M.; Fletcher, T.; White, L.; Jonsson, C. Development, validation and optimization of a luminescence-based high throughput screen for inhibitors of Severe Acute Respiratory Syndrome. J. Biomol. Scr. 2007, 12, 33–40.
- Noah, J.; Severson, W.; Noah, D.; Rasmussen, L.; White, E.L.; Jonsson, C.B. A cell based luminescence assay is effective for high throughput screening of potential influenza antivirals. Antiviral Res. 2007, 73, 50–9.
Dr. Jonsson is Program Leader, Emerging Infectious Diseases
Research Program; Ms. White is Manager, High Throughput
Screening Center and Enzymology Laboratory; Dr. Noah is
Research Biologist; Dr. Severson is Research Biologist; and
Dr. Heil is Associate Research Biologist, Emerging Infectious
Diseases Research Program, Southern Research Institute, 2000
9th Ave. S., Birmingham, AL 35205, U.S.A.; tel.: 205-581-2681; fax: 205-581-2093; e-mail: [email protected].