Magnetic particle-based techniques are widely used in many diverse biological applications, offering a relatively inexpensive technology and subjecting samples to very little mechanical stress. Although referred to as “magnetic,” the majority of the particles used are paramagnetic or superparamagnetic, meaning that the beads only exhibit magnetic properties in the presence of a magnetic field, with no residual magnetism once removed. A wide range of bioreactive molecules can be adsorbed or coupled to the bead polymer surface and used in the separation of biological materials such as cells, proteins, DNA, and RNA. Paramagnetic beads are particularly suitable for automated procedures because the instrumentation exists to easily mix, incubate, and separate the particles in 96-well plates without columns or centrifugation. The technology uses magnetic rods to transfer particles through the various purification phases of binding, mixing, washing, and elution, rather than moving solutions around.
Increasing range of applications
Magnetic particles are increasingly being used as carriers for binding proteins and enzymes in proteomics applications, where the immobilized biomolecules can be used directly for a bioassay or as affinity ligands to capture or modify target molecules or cells. In addition to whole cell isolation, even cell organelles can be selectively separated using magnetic particles. Immunomagnetic cell isolation and separation methods are also proving useful in cell sorting applications, for example, in the isolation of rare progenitor cells from human cord blood.
Magnetic particle separation has proven most invaluable, however, for sample preparation in drug discovery and genomics applications, including high-throughput genome isolation for sequencing or PCR amplification for downstream processing such as single nucleotide polymorphism (SNP) analysis or expression profiling. The particles offer many advantages, including reduced reagent costs, simplified procedures, and reduced time needed to achieve good yields of high-purity nucleic acids.
Flexible solutions for nucleic acid purification
Magnetic particle-based kits offer excellent performance for the purification of DNA and RNA. The range of starting material for nucleic acid research can be broad—from 5 mL of blood for routine testing to very small samples, such as those obtained from forensic surface and contact swabs, hair, and cigarette butts. The technique enables the isolation of DNA or RNA in sufficient quantities from all kinds of material sources such as serum, blood, bacteria, cell culture, tissue, food, soil, or feces. Kits can also be used to purify DNA from several plant species and sources. Use of magnetic bead-based separation technology ensures that the resulting DNA and RNA are free of protein, nucleases, and other contaminants, ensuring that purified nucleic acid samples are ready to use in downstream applications.
Speed and sample purity continue to be a major concern to genomics and proteomics researchers. The sheer volume of work involved in genome and protein mapping means that processing speed is essential. Unsatisfactory results on even the most sophisticated analyzers are increasingly linked to poor initial sample quality. The variety of samples being used, however—from rare cell types and mRNA transcripts to increasingly large and delicate protein complexes—presents a real challenge. Automating such purification procedures ensures impurities are removed rapidly, efficiently, and consistently; an open and flexible automation system will enable the user to choose any available magnetic particle-based purification kit suitable for the application.

Figure 1 - KingFisher Flex magnetic particle processor.
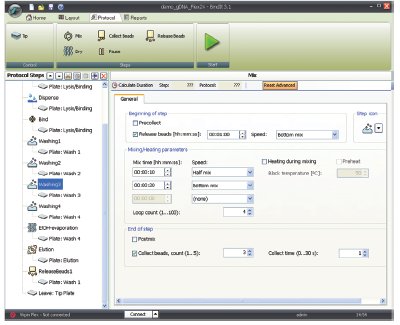
Figure 2 - BindIt 3.1 software parameters.
Predefined application protocols are available for many existing magnetic particle-based methods, and it is easy to customize existing protocols or create entirely new ones using appropriate intuitive software. In addition, it is important to select a flexible processor that offers high throughput and a wide range of processing volumes, such as the KingFisher® Flex (Thermo Fisher Scientific, Vantaa, Finland) (Figure 1). The purification protocol is created with the associated Thermo Scientific BindIt 3.1 software, which allows the user to optimize the purification conditions taking into consideration the sample type and particles used in the extraction kit (Figure 2). Several different magnetic heads and plate formats are available. This study used 96-deep-well and 24-well magnet heads and tip combs.
Optimization of the DNA purification protocol
The DNA purification protocol typically consists of cell lysis/DNA binding, several washing steps, and the DNA elution. All of these purification steps were optimized for both 96- and 24-deep-well formats. In the 96-well format, the DNA extraction was performed from 200 μL blood or ~6 μg pure calf thymus DNA, and in the 24-well format, from 1 mL blood or ~35 μg calf thymus DNA. Blood DNA purification kits from two vendors were used. The absorbances of the eluents were measured, the DNA yield was calculated based on the measured 260-nm absorbance, and the quality was determined by analyzing the 260/280 nm ratio. The quality of DNA was also tested by end-point polymerase chain reaction (PCR) to control the presence of PCR inhibitors in the eluent. The protocols were optimized by changing one variable at a time, e.g., the mixing speed of one step was optimized by alternating the available mixing speeds while the rest of the protocol remained unchangeable.
Effects of different mixing speeds
Table 1 - Optimizing binding and elution of the DNA purification protocol

The different mixing speeds had a strong effect on the DNA purification. For DNA purification, it is important to mix the solutions efficiently to reach optimal binding of the DNA and the bead. The faster mixing speeds increased DNA yields for DNA purification from blood. When one variable at a time (binding, washing, and elution) was tested, the faster mixing speeds for all steps resulted in a better yield (Table 1). The mixing speed makes the greatest difference to the results with the 24-well format, due to the shape of the tip and the well. Powerful washing is essential for the high purity of the DNA. The protocol parameters in the binding and elution steps affected, primarily, the total DNA yield.
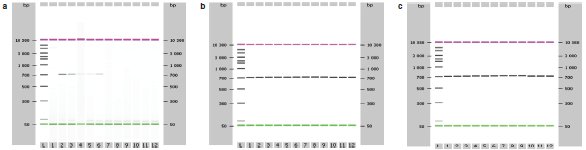
Figure 3 - Effects of the wash step mixing speed on the final DNA quality. PCR products of the elutions. a)Slow, b) medium, c) fast.
In the washing steps, the slow mixing had an effect on the DNA quality. The purified DNA was analyzed with PCR, and the results show the remaining impurities affecting the secondary applications (Figure 3). Table 1 shows the effects of different mixing speeds for binding and elution steps and the effect of the heating and elution volume on the final DNA yield. Results of the slow mixing speed indicate the impurities in the elution and inhibition of the enzymatic reaction due to inefficient washes. Usually, slower mixing speeds are preferred during elution due to DNA degradation, but, according to this study, fast mixing in the elution results in the best quantity and quality DNA. The slow mixing speed is useful for the heated steps of the nucleic acid purification.
Table 2 - BindIt 3.1 software mixing speed recommendations for DNA purification
with KingFisher Flex

Table 2 recommends the optimum mixing speeds for DNA purification in this setup. It should be noted, however, that the same optimization is not directly applicable to RNA and protein purification, for example. Half mix, medium, or slow mixing speeds are recommended for more sensitive bead–biomolecule complexes.
Conclusion
Magnetic separation technology is increasingly becoming an integral part of the laboratory in this postgenomic era, finding a role in biomedical, clinical, and cell sorting applications and proving an invaluable part of the process in proteomics, drug discovery, and genomics. The range of magnetic particles now available enables routine separation and purification of biological materials such as cells, proteins, DNA, and RNA. Automation of this technology offers many advantages in terms of increased efficiency, throughput, reliability, consistency, reduced costs, and enhanced yields. All of the purification steps require beads to separate effectively, and this study found that mixing speeds are significant for optimal binding, efficient washing, and active elution. By simple optimization of the magnetic particle-based nucleic acid purification protocol, it is possible to provide a rapid and reliable method for DNA isolation and achieve a good yield of high-quality DNA.
Use of an open-platform automation system and intuitive software, which is easy to modify, enables the user to take into consideration differing sample types and particles. Rapid and reliable separation methods and easy adjustment of isolation protocols should allow complete flexibility of use for all future applications.
The authors are with Thermo Fisher Scientific, Ratastie 2, P.O. Box 100, Vantaa, FIN-01621, Finland; tel.: +358 9 32910200; e-mail: [email protected].