Science-driven decisions depend on data, both structured (in tables) and from notes (natural language). Data may reside in disparate databases, in electronic data files, or even be printed, and file and database architecture, nomenclature and column/row labels often evolve over time.
The evolving technology landscape makes change inevitable, and can create problems. For example, an FDA audit might ask, “How does a drug shipped today compare with the product licensed 20 years ago? Does it show the same characteristics for purity and stability? Answering can require days or even months of searching unsupported legacy files.
To be useful, “big data” must be harmonized, connected and searchable. With this as background, editor emeritus Robert L. Stevenson interviewed Robert Stanley, co-founder and President of IO Informatics (Berkeley, Calif ). IOI has focused on integrating diverse scientific files and databases. Data harmonization and integration are core capabilities of IOI’s Sentient software platform.
RLS: How did you get started in data integration?
RS: In 2002 Dr. Erich Gombocz (an IOI co-founder) and I were aware that more effective informatics tools were needed. We were familiar with the strengths and limitations of relational databases (RDBs). We recognized that integration and interoperability were weak links. Researchers struggled with disconnected data silos. The human genome had been sequenced, with more genomes in the pipeline, but research suffered from inadequate linking of genetic to epigenetic information. We envisioned semantic technologies (ST1) promoted by the World Wide Web Consortium (W3C2), which developed the Resource Description Framework (RDF3) for data integration, and had confidence that ST would address critical difficulties in describing, integrating and searching disparate data.
We’ve confirmed that using RDF with the right support tools makes it possible (for) integration experts (we [IOI] call them “knowledge engineers”) to solve the toughest healthcare/life science (HCLS) integration challenges. IOI has helped many top pharma firms save time and money. We provide software and methods required to integrate huge data resources. Scientists and business users can easily query integrated resources without ongoing support from IT departments. We have published case studies where IOI has solved challenging integration problems where bigger, more widely known companies have failed.
RLS: Why is data integration an issue today?
RS: Discoveries are blocked if an answer is not available from accessible data. In data-driven research, scientists should be able to ask detailed questions and get results quickly, with precision and accuracy.
Often scientists suspect that answers exist in combinations of several files or databases, but accessing them requires time and skills that they do not have. Implementing a query requires help from a specialized data scientist or relational database expert with advanced knowledge not related to the research.
Conventional RDBs suffer big challenges with the extract/transform/load (ETL4) process for data harmonization and integration. Turnaround times for conventional integration are often measured in years for enterprise research resources. Workarounds include repeating experiments that were done before, or simply avoiding the question. Often an IT department finally provides a resource, but the research (sic) has already moved on.
It’s surprising, but many leading companies still use teams of experts for manual curation, piecing data together for each new research initiative. Expensive teams and months of time are wasted tracking down and putting together data when technologies like ours can solve these problems efficiently at the enterprise research level.
RLS: How does ST facilitate data integration?
RS: ST employs universally descriptive subject-predicate-object “triples” (e.g., “gene-expresses-protein” is a triple) to define data relationships. In contrast to the rigidity and obscurity of RDB tables, using triples as basic assertions to link data facilitates agile and meaningful integration. Triples combine to create rich, domain-specific data models or ontologies (see Figure 1).
 Figure 1 – RDF assertions combine to create “graphs” which are data connected and organized by ontologies or data models for formal reasoning or for less application uses.
Figure 1 – RDF assertions combine to create “graphs” which are data connected and organized by ontologies or data models for formal reasoning or for less application uses.The identity of each data instance, including its relationships (even if they come from different data resources), is attached to the data. This simplifies integration for precise searches by relying on comprehensive, explicitly meaningful connections to other integrated resources.
Application of triples and ontologies rather than database schemas brings benefits to data modeling, integration and search. For example, ST makes it possible for automated inference to manage translation and encoding of coded and free-form values. Translation is supported by visual oversight, with provenance kept for changes, and alerts for problems with source data.
We’re working with a clinical institute that performs neurodegenerative disease research. Over the past 20 years, the institute has applied a variety of systems to collect data. While defining project goals, research leaders asked, “Could it be possible that our rich existing data describes diseases and cures?” The consensus response was, “Probably yes, for some cases. It’s hard to know since much of the data is not readily available as it is.”
We applied the Sentient platform to integrate data from multiple sources (files, instruments, databases, public sources) without creating new isolated silos. This involved translation into standardized nomenclature and linking using a “patient-centric” ontology under the open W3C standard, RDF. We connected content from databases, files, instruments and applications regardless of format and structure. This created an extraordinarily comprehensive knowledge base that is updated automatically.
The first step was to use the Knowledge Explorer component of the Sentient platform to identify, extract and transform inconsistent data in each DB (see Figure 2). Once the DB is consistent, it is easy to load the data into unified RDF. The first phase alone provided researchers with the ability to easily search hundreds of man-years of integrated data for the first time. The integration (extraction and transformation stage) was accomplished in under a year.
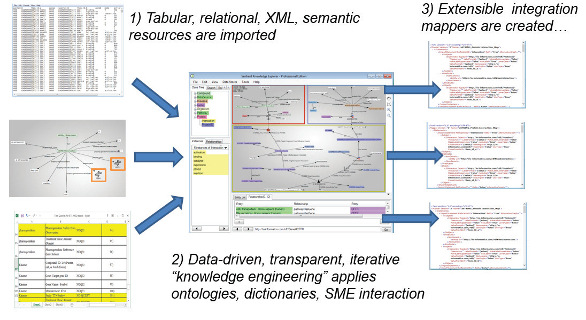
F
igure 2 – Extraction and transformation parts of ETL. 1) Data from structured files or text mining output (upper left), semantic resources (left center) and databases (bottom left) are identified and imported. These are transformed into stores of linked data or “triples,” which enables 2) data harmonization and integration with the aid of lexical matching, domain-specific ontologies and inference made possible by the semantic data modeling and visualization tool (“Knowledge Explorer” center). 3) This advanced visual query and inference engine generates data harmonization and integration rules (right) and can also be directly applied to explore the integrated data for research and discovery. The integrated data is prepared for delivery to third-party tools and can also be searched with advanced semantic query languages such as SPARQL.RLS: You mentioned provenance. What about keeping track of changes made to data?
RS: Provenance is critical for valid research. It is important to keep track of where data comes from, along with any transformations. We’ve worked with AstraZeneca to clean and integrate data gathered during thousands of clinical trials. Numerous integration decisions needed to be made. For example, thousands of column names may have included “nanomolar,” “NM” or “nm”—were all of these nanomolar? Were there nanomaterials? LC50 values? Were these wavelengths or could the column headers “nm” indicate patient name? Not measured? Manual review and curation of this data could have taken years.
This is where ST’s ability to apply automated inference can be very helpful. A trained integration expert (or knowledge engineer) can create innovative algorithms using RDF-based query and inference methods to analyze data context. Context removes ambiguity and allows the system to transform or add assertions to an existing dataset, called “entailment.”5
IOI’s system makes it possible to automatically apply entailment to generate preferred names and connections, with tracking of changes made. Availability of domain ontologies and vocabularies that describe how data should be named and linked according to common standards is also useful. IOI has connections to leaders in the ontology space, including Stanford’s National Center for Biomedical Ontologies (NCBO). We have direct connections from our software to resources made available by NCBO and to other Linked Open Data (LOD) resources available online.
It’s possible for a novice to deal with this sort of complexity by trying to sort of link everything to everything. Sure, you can run queries in that environment, but you will see poor performance with results that you probably won’t be interested in. IOI is happy to teach customers the basics, preferably as part of a project. To ensure useful outcomes we hope customers will keep us around for advanced support when needed. Either way, connecting data is much easier with RDF than traditional restructuring or joining of files is within RDB schema.
RLS: Okay, what about big data? Integrating large data sets?
RS: The ability for ST to handle big data has turned the corner. Databases are now handling over 1 trillion triples with good performance.6 IOI applies best-in-breed high-performance computing, working with Oracle, Franz, OpenLink, Cray Computers, Neo4j, MongoDB and others. Our platform architecture supports “infinite horizontal scalability” via the cloud. We can host or install high-performance software on systems in-house. We’re on an exponential curve for working with big data at this point.
We’re seeing massive growth in scale on our production pharma systems. We’ve been able to handle this growth gracefully. I’m particularly excited about [the] performance we’re seeing from beta evaluators of IOI’s pending release of the Sentient Enterprise Platform solution, which is scheduled for a Q4 2015 launch. This platform performs well with relatively low-cost server hardware, and is horizontally scalable for enterprise use on massive internal or external server clouds as needed.
Scientists can construct queries using convenient filters or browsing methods, specify data sources, and get results in seconds. Pattern-based searches can be run applying automated reasoning across global datasets. Queries that apply advanced inference on big data may demand comparably powerful server processing or they may take some time to complete. Time required to ask questions decreases and the range and quality of questions and answers improves.
RLS: Is integrating data worth the effort?
RS: Absolutely! Integration of complex data has been difficult with older technologies. Efficient integration using ST is now routine and provides several new immediate and long-term benefits to HCLS research today.
If one does not have clean, integrated data, the platform will give poor results. If integration is done well, data can be used over and over again and extended as needed for growing decisive knowledge. Research costs are reduced by orders of magnitude.
ST also delivers improved efficiency in long-term enterprise data management and integration. RDF is not a proprietary standard. Similar to document description standards like HTML, RDF complies with the global W3C standard for data description. Semantic data is usefully “open” and prepared for interoperability with new data, to support new questions and resources as they are needed. Integrated data becomes more accessible over time when using RDF.
Clinical research is a great example for integration being worth the effort. For example, PROOF Centre at the University of British Columbia’s St. Paul’s Hospital is one of IOI’s early success stories. PROOF sought to expand their biomarker program for autoimmune diseases. Internal integration was effective but constrained in scope. Integrating external databases for reference information was problematic. Searches were costly.
The Sentient platform delivered unified reporting, analytics and resulting applications with a scope ranging from biomarker discovery to precision medicine applications (see Figure 3). Data from diseases, sample banks, clinical results, treatments, genomics, proteomics and metabolomics was integrated and could be directly interrogated. Patterns that combine multiple blood-based biomarkers to detect patients at risk of autoimmune events (without requiring costly biopsies that are commonly taken) have been discovered and are in FDA review. Having this data at hand reduces the need for costly new studies.
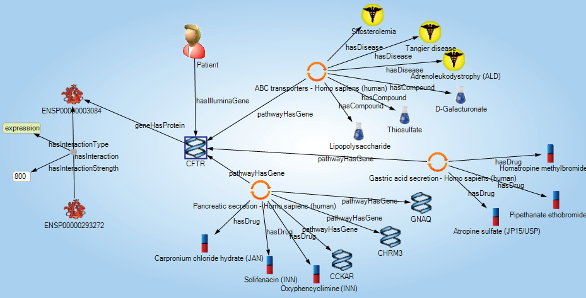 Figure 3 – Patients are at the center of a semantic network that connects samples, assays, treatments and disease endpoints.
Figure 3 – Patients are at the center of a semantic network that connects samples, assays, treatments and disease endpoints.Bruce McManus, M.D., Ph.D., managing director of PROOF and a member of IOI’s Scientific Advisory Board, finds that the ability to consume and intuitively represent a wide variety of data types including images, text and numerical is at hand. Further, we are now able to display the data in ways that make significant features immediately obvious to our biologist end users. These features enabled our research to move to a completely new level.
References
- www.w3.org/standards/semanticweb/
- www.w3.org/
- www.w3.org/RDF/
- https://en.wikipedia.org/wiki/Extract,_transform,_load
- https://en.wikipedia.org/wiki/Logical_consequence
- http://download.oracle.com/otndocs/tech/semantic_web/pdf/OracleSpatialGraph_RDFgraph_1_trillion_Benchmark.pdf
Robert L. Stevenson, Ph.D., is Editor Emeritus, American Laboratory/ Labcompare; e-mail: [email protected]