In 2007, researchers created the
first human induced pluripotent
stem cells (iPS cells) by infecting
skin cells with four transcription
factors (Oct-4, Klf4, Sox-2, and
c-Myc).1 After the cells had achieved
an induced pluripotent state, the
researchers were able to differentiate
them into new cell types. The ability
to revert somatic cells to an embryonic
state and then differentiate them
into desired lineages offers a wealth of
opportunities for disease research.
In addition to their potential as therapies
based on cell replenishment, iPS
cells are a novel tool for in vitro disease
modeling and drug screening. As
more and more researchers incorporate
iPS cells into their studies, the technology
to create and characterize them
continues to be refined while new
questions and challenges are revealed.
Evolving technology
Initial efforts to generate iPS cells
required simultaneous coinfection
of cells with four separate retroviral
expression vectors. Each vector carried
one transcription factor, which resulted
in a high number of genomic integrations.
Alternative approaches to iPS generation
have included use of plasmids and nonintegrating
adenovirus vectors to deliver
the transcription factors. The rates at which
cells convert to pluripotency using these
methods, however, are far lower than those
obtained using retroviral vectors.2
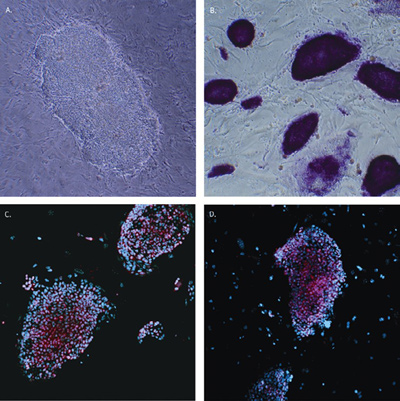
Figure 1 - Mouse iPS cell morphology and marker expression. Mouse
embryonic fibroblasts (MEFs) infected with the STEMCCA lentivirus
display characteristic ES cell morphology in a phase contrast
image of a single mouse iPS cell colony seven days after infection
(a). Passage 3 mouse iPS cells exhibited high alkaline phosphatase
activity (b) and express high levels of Oct-4 (c), SOX-2 (d), and
SSEA-1 (not shown). Cell nuclei were counterstained with 4ʹ,6-diamidino-2-phenylindole (DAPI) (blue).
Generation of human and mouse iPS
cells can now be accomplished using a
single, excisable polycistronic lentiviral
vector that delivers all four Yamanaka
transcription factors (STEMCCA™,
EMD Millipore
[Billerica, MA]; Figure
1). Use of a single vector significantly
reduces the number of viral integrations
required—in some cases, iPS clones possessing
only a single viral integrant can
be isolated.3
In addition to fully reprogrammed cells,
transcription factor-induced reprogramming
can generate undifferentiated cells
that are not completely pluripotent.1,4
These partially reprogrammed cells (pre-iPS
or Class I iPS cells) have global gene
expression and DNA methylation patterns
distinct from embryonic stem (ES) cells,
despite similarities in colony morphology
and the ability to propagate extensively in
culture. Partially reprogrammed cells can be
characterized by: continued expression of
the viral transgenes; incomplete expression
of pluripotent genes such as Nanog, SSEA-4, and TRA-1-60 with human cells; down-regulation
of somatic cell marker genes;
and in mouse cells, the inability to generate
germ line transmitting chimeric mice.
A range of kits and reagents are available to
assist researchers in their characterization of
iPS cell cultures. Access to optimized tools
and protocols can help accelerate research
as more laboratories that are new to the stem
cell field begin to work with iPS cells.
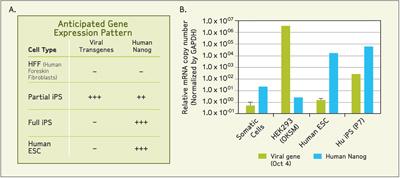
Figure 2 - a) Anticipated gene expression pattern of viral transgenes and Nanog, a marker of pluripotency for
different cell types. b) Relative mRNA copy numbers for viral and Nanog genes normalized using glyceraldehyde
3-phosphate dehydrogenase (GAPDH) expression levels. Relative mRNA copy numbers of viral and Nanog genes
from human iPS cells were plotted together with those from negative control (somatic cells) and positive control for
viral gene (HEK293 cells transfected with OKSM viral vector) and human Nanog gene (Hu ESC).
Assessment of whether an iPS clone is a fully
or partially reprogrammed colony can be rapidly
accomplished using a multiplex real-time reverse transcription-polymerase chain reaction (RT-PCR) kit (STEMCCA
Viral Gene Detection, EMD Millipore).
The kit detects the expression of
the STEMCCA viral transgenes and ES/iPS cell pluripotent markers. Because the
STEMCCA lentivirus is a single polycistronic
vector containing four transcription
factors, all the transgenes are
transcribed as a single mRNA molecule.
Subsequent transgene analysis shows the
gene expression level of the entire cassette
(Figure 2). In a fully reprogrammed
colony, one would expect to see the
down-regulation of viral transgene expression
and the up-regulation
of Nanog gene
expression, a pluripotency marker.
A rapid, efficient flow cytometry-based
process originally developed to characterize
human ES cells can also be used
to evaluate the expression of markers
reflecting the degree of pluripotency of
iPS cell lines. The FlowCellect hESC
Characterization Kit (EMD Millipore)
contains validated fluorescent antibodies
to Oct-4; SSEA-4; and SSEA-1,
a marker for monitoring transition of
undifferentiated stem cells to a differentiated
state. The labeled antibodies
are optimized for cytometric analysis
on the guava EasyCyte™ benchtop flow
cytometer (EMD Millipore).
Typical cytometric results show iPS cell cultures
having a distinct Oct-4 and SSEA-1
positive population of cells with significant
separation from parent fibroblasts. Pluripotency
is indicated by a shift in fluorescence
intensity resulting from an increase in the
expression of Oct-4 and SSEA-4, which are
markers of acquired pluripotency. In contrast,
the population of iPS cells expressing
SSEA-1, a marker that coincides with
human embryonic stem cell differentiation,
is reduced. This same cytometric technique
can also be used to compare different clones
of the same iPS cell line.
Most recent efforts to reprogram human
somatic cells to iPS cells utilize synthetic
mRNAs encoding the four Yamanaka factors, which overcomes the problems associated
with genomic integration and insertional
mutagenesis.5
Disease modeling
iPS cells provide the ability to recapitulate
both normal and pathological tissue formation
in vitro, and can yield a genetically
diverse set of patient- and disease-specific
cells. Following well-established protocols, a
disease researcher can take a skin biopsy from
a patient and reprogram the isolated fibroblasts
into iPS cells. Those iPS cells can then
be differentiated into the cell types that are
affected in the disease being studied, given
the appropriate culture conditions.
A barrier to realizing the full potential of iPS
cells is the ability to direct their differentiation
into the cell types of interest. Identifying
the right cocktail of media conditions, supplements,
and growth factors that successfully
drive iPS cells toward a desired lineage
on a reproducible basis is a time-consuming,
iterative exercise. A carefully choreographed
series of signals must be recreated to guide
cells down the chosen pathway. This labor-intensive
work has already been done for a
number of cell types. Kits and media containing
an optimized set of factors necessary
to differentiate stem cells to a chosen lineage
are commercially available for generating
neurons, oligodendrocytes, mesenchymal
cells, and osteocytes.
iPS technology enables researchers to culture
cell types that would normally be a challenge
to access, such as those affected by neurological
disorders. Diseases with long latency
periods such as Alzheimer’s, ALS, or Parkinson’s,
however, may prove difficult to model
in relatively short-term in vitro cultures. It
may be unrealistic to expect a phenotype to be
revealed after a few weeks in culture of a late
onset disease that takes decades to appear in
patients. An additional complicating factor is
that cells derived from iPS cells are more fetal
in nature, reflecting a young developmental
stage. External factors mimicking some type of
environmental stress may be required to speed
the in vitro aging process.
The true power of iPS technology is the
potential to create multiple cell types from
a single patient that are believed to interact
in the development and progression of complex
diseases. As multiple cell lineages are
incorporated into disease models, researchers
are exploring ways to recreate the three-dimensional
in vivo setting for these models.
Advances in tissue engineering will
enable multiple cell types to interact and
communicate with each other in a manner
that more closely resembles their in vivo
environment. Incorporation of advanced
scaffolds and matrices may reveal phenotypes
that are not obvious with single cell
types or even multiple cell types co-cultured
in a two-dimensional environment.
Understanding the microenvironment that
iPS derivative cells may face when they are
transplanted back into the body is also critical.
Dr. Aileen Anderson and her team at the
University of California at Irvine are exploring
whether neurons derived from fetal neural
stem cells, embryonic stem cells, and iPS cells
can be used to mediate repair in spinal cord
injuries. Their focus is to understand what
the role of the inflammatory microenvironment
will be in dictating how a cell population
responds after transplantation. The
team is studying how cells from these different
populations are going to be influenced after
transplantation in terms of their fate, their
migration, and how the environment they see
is going to signal back to those cells.
Drug screening
iPS derivative cells offer unique advantages
when incorporated into the traditional
small-molecule drug discovery and development
process. Disease-specific iPS derivative
cells can be used to assess and optimize
lead compounds and facilitate metabolic
profiling and toxicity evaluation.
Using iPS derivative cells, potential therapeutics
can be screened against a large
number of patient-specific cells prior to
initiating clinical trials. Variation in the
response to drugs by cells of patients with
genetic differences can guide more targeted
selection of patients for enrollment
in clinical trials, resulting in trials that are
smaller and more likely to be successful.
Dr. James Ellis, senior scientist at the Hospital
for Sick Children in Toronto and co-scientific
director of the Ontario Human iPS
Cell Facility, is interested in drug screening
applications for cystic fibrosis. He and his
group are looking to identify reproducible
phenotypes first. Once this is done, high-throughput
screening will be a very effective
way to identify small molecules that may be
effective for a particular patient.
Dr. Ellis readily sees the value in the derivation
of lung cells from iPS cells. According
to him, “Obtaining lung cells from a patient
with cystic fibrosis is really only possible
when they’ve undergone a lung transplant.
But one of the consequences of the disease
is that patients have dramatic lung infections
so it’s very difficult to establish primary
cell lines and even if you did, those
will have a limited ability to be passaged.
You may not be able to make enough cells
to complete or verify your screen.”
Through use of iPS cells, the laboratory
plans to generate large numbers of cells
from a range of patients. Genomic patterns can then be cross-referenced to drug
screening results. The researchers can then
compare one patient to others and possibly
start to make predictions as to which drugs
are going to work in which patients.
Investigative toxicity
A significant advantage of using iPS cells
in drug screening applications is the ability
to conduct toxicity tests on cells of the
same patient. If a drug is found to be efficacious,
it can be tested against cardiomyocytes
and hepatocytes derived from the
same iPS cells. Use of iPS cells for toxicity
testing can also help overcome the inherent
limitations of current methods.
The liver plays a central role in processing
and metabolizing drugs and other substances
in the bloodstream. Because hepatocytes
are responsible for metabolizing most compounds
that enter the body, these cells are
used during the drug discovery process to predict
how drugs will be metabolized and to
what extent a drug may be toxic to the liver.
Drug-induced liver injury is the principal
reason clinical trials are suspended
and approved drugs withdrawn from the
market. In fact, drug-induced liver injury
has been the most frequent, single cause
of safety-related withdrawals of marketed
drugs over the past 50 years.6
Investigative in vitro liver toxicity studies
are typically conducted using primary
human hepatocytes or an immortalized
(genetically transformed) liver-derived
cell line such as HepG2. Despite routine
use for investigative toxicity, both of these
options present significant drawbacks:
-
Primary human hepatocytes are derived
from fresh liver tissue, which is typically
sourced from cadavers or cancer resections.
Supplies can be limited and the
tissue can vary widely among donors.
- Primary hepatocytes cannot be sustained
for more than a few days in culture
without losing function.
- Although immortalized hepatocyte cell
lines can be cultured indefinitely, these
cells display distinct differences from
normal liver cells and may not exhibit
normal cell behavior or response.
The challenges of hepatocyte-based in
vitro toxicity testing have led drug developers
to rely heavily on animal models for
preclinical metabolism and toxicity testing.
But animal models may not be fully
and reliably predictive of human toxicity,
are low throughput and expensive, and
raise ethical concerns for some.
Cost and throughput often relegate use of
animal models to the later stages of preclinical
development, after a company has
invested significant resources and time in
a lead compound. This delayed evaluation
of toxicity contributes to the high
failure rate of compounds in late-stage
preclinical testing, which is extremely
costly. Earlier, more effective assessment
of drug candidate toxicity has the potential
to reduce the attrition rate of drugs in
later stages of development.
Differentiation and expansion of human
iPS cells into functional hepatocytes for use
in investigative toxicity studies could overcome
the shortcomings of primary hepatocytes
and immortalized cell lines. Use of
iPS-derived hepatocytes (and other cell
types commonly used for toxicity studies)
offers a number of important advantages to
investigative toxicity studies, including:
-
Availability of a consistent source of
cells that more closely match in vivo
phenotype and physiology
- Elimination of reliance on donor sources
that can have sporadic availability
- Reduction in the use of animal models
and animal tissue
- A more standardized, reproducible process
for toxicity testing
- Improvement in the predictive capabilities
of early toxicity studies, leading to
reduction in late-stage attrition of drugs.
More efficient and predictive toxicity studies
enabled by iPS-derived cells can be expected
to reduce development costs associated with
the late-stage failure of drug candidates.
Identifying drug candidates with toxicity
concerns earlier in the discovery process can
improve the safety and, ultimately, the successful
outcomes of clinical trials.
The ability to differentiate iPS cells into
a wide range of lineages has created new
opportunities in both clinical and research
settings. Use of these cells for disease modeling
and drug and toxicity screening can
help overcome the limitations of current
methods, enable construction of human
models of complex diseases, and reveal
important insights that can lead to a more
personalized approach to medicine. Alongside
their potential to reshape the research
and clinical landscape, these remarkable
cells have presented researchers with new
questions to explore, challenges to address,
and applications to discover.
References
-
Takahashi, K.; Tanabe, K. et al. Induction
of pluripotent stem cells from adult human
fibroblasts by defined factor. Cell2007,
131, 861–72.
- Baker, M. Integration-free iPS cells. Nature
Reports Stem Cells; Oct 16, 2008.
- Sommer, C.A.; Gianotti Sommer, A. et
al. Excision of reprogramming transgenes
improves differentiation potential of iPS
cells generated with a single excisable vector.
Stem Cells2010, 28(1), 64–74.
- Jaenisch, R.; Young, R. Stem cells, the
molecular circuitry of pluripotency and
nuclear reprogramming. Cell 2008, 132(4),
567–82.
- Warren, L.; Manos, P.D. et al. Highly efficient
reprogramming to pluripotency and
directed differentiation of human cells
with synthetic modified mRNA. Cell Stem
Cell2010, 7(5),618–30. In press.
- Guidance for Industry: Drug-Induced Liver
Injury; U.S. Department of Health and
Human Services, July 2009.
Dr. Chu is R&D Manager, and Ms. Noble and
Ms. Rollins are Product Managers, Stem Cells/Cell Biology, EMD Millipore, 290 Concord Rd.,
Billerica, MA 01821-3405, U.S.A.; tel.: 781-533-6000; e-mail: [email protected].