Reduced costs and higher throughput have made next-generation sequencing (NGS) more accessible to users in such diverse fields as diagnostics, infectious disease, food safety and public health. NGS has revolutionized these areas as a result of its ability to produce genomes on an unprecedented scale1-4 in less time and at a lower cost. Increased capacity has created a challenge: establishment of an automated library preparation workflow to enable reliable and robust sequencing.
For every application of NGS there is a specific protocol to convert the source nucleic acid to a standard DNA library. The basic NGS library preparation procedure involves nucleic acid isolation, adapter and bar-code ligation, DNA fragment size selection, amplification and amplification reaction cleanup.
Magnetic beads play a significant role in many of these steps, highlighting the need for their uniform distribution (Table 1). An efficient bead mixer placed on the deck of a robotic workstation can potentially keep the beads uniformly suspended in the source tube or reservoir before being distributed to the target wells of the library preparation plate.5 Since the beads are precious and supplied in small volume, the mixer is expected to minimize dead volume. In addition, bead mixers provide advantages over pipet mixing, as the latter results in the beads clumping or sticking inside the pipet tips, which leads to inefficient mixing and nonuniform distribution of the beads onto the target plate.
Table 1 – Applications of magnetic beads in next-generation sequencing

The study presented in this article demonstrates use of a bead mixer installed on the VERSA 1100 NGS library preparation workstation (Aurora Biomed, Vancouver, B.C.).
Materials and methods
Determination of uniform bead distribution
The materials and methods from Gill et al.5 were used to validate a bead mixer installed on the VERSA 1100 workstation. The magnetic beads were suspended in 5 mL of lysis solution used for nucleic acid isolation; the tube containing the beads was inserted into the bead mixer. Next, using the system’s single-channel functionality, 20 μL of beads was automatically distributed from the bead mixer to each well of a 96-well multiple-well plate. The sampling was carried out in replicates of n = 10 to check the uniform distribution of the beads. The beads from each well of the target plate were resuspended in 1 mL of distilled water to read OD600 on the Spectrumlab 22PC spectrophotometer (Spectrumlab, Shanghai, China).
Automated NGS library preparation
The VERSA 1100 NGS automated liquid handling library preparation workstation was further evaluated for its ability to produce high-quality, consistent libraries for NGS on the Ion Torrent PGM (Life Technologies, Grand Island, N.Y.). In the NGS library preparation protocol, the bead mixer was used for cleanup procedures for both the unamplified and amplified libraries, including first-round purification, second-round purification and fragment size selection as per the manufacturer’s protocol. Agencourt AMPure XP magnetic beads (Beckman Coulter Inc., Brea, Calif.) were used for cleanup. The Ion AmpliSeq library preparation kit and Ion AmpliSeq ready‐to‐use cancer panel kit (Life Technologies Inc.) screened the loci listed for allelic frequency associated with cancer hot spots. The Ion PGM sequencer (Life Technologies) was used to generate the NGS data.
Results and discussion
Automated bead distribution and consistent bead delivery
Previous validation studies on installation of the bead mixer on the VERSA 1100 NGS library preparation workstation were encouraging.5 This process, as shown in Figure 1, was validated for distribution of the magnetic beads prior to its application for AMPure XP beads for NGS library preparation. Figure 2 shows a standard curve that was generated with the OD600 values from a spectrophotometer obtained with manually mixed bead suspensions. The bead concentrations delivered by the VERSA liquid handling workstation were uniform, consistent and reproducible (Figure 3).
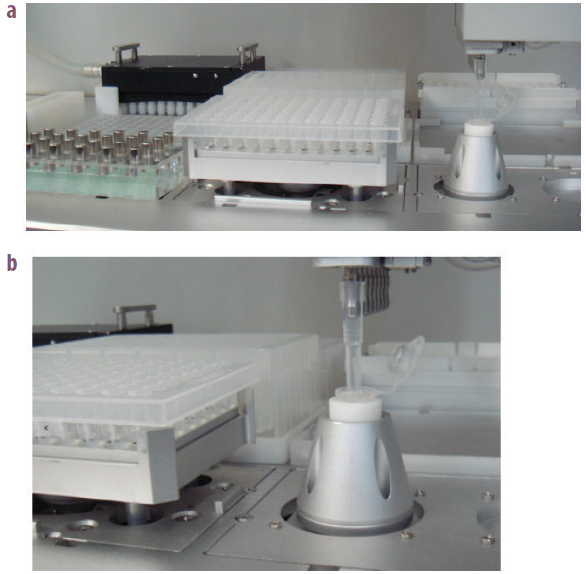 Figure 1 – Bead mixer installed on the deck of VERSA 1100 NGS library preparation workstation.
Figure 1 – Bead mixer installed on the deck of VERSA 1100 NGS library preparation workstation.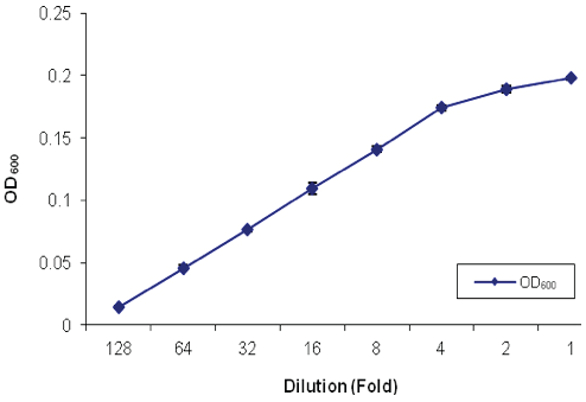 Figure 2 – Standard curve of the magnetic bead suspension prepared by manual suspension (n = 3).
Figure 2 – Standard curve of the magnetic bead suspension prepared by manual suspension (n = 3).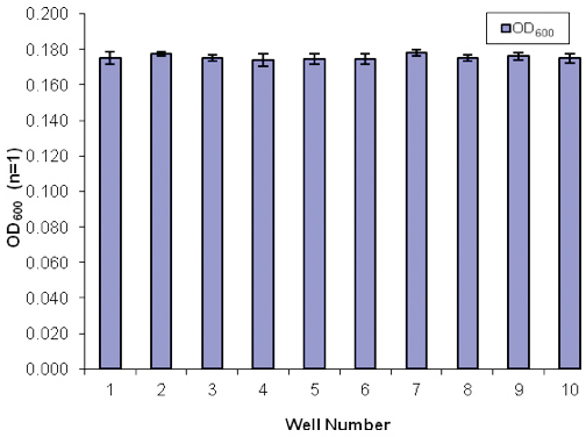 Figure 3 – Automated distribution of the magnetic bead suspension (automated mixing) to the target well of the multiple-well plate for nucleic extraction or purification (n = 3).
Figure 3 – Automated distribution of the magnetic bead suspension (automated mixing) to the target well of the multiple-well plate for nucleic extraction or purification (n = 3).Library preparation on the VERSA 1100
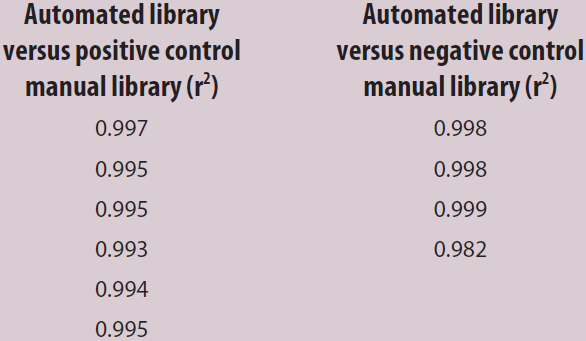
NGS is of great benefit in cancer diagnostics: it enables practitioners to examine specific sets of single-nucleotide polymorphisms (SNPs) and genes associated with patient tumor biopsies that have established relevancy to a particular cancer phenotype. It also aids in diagnosis and helps guide treatment.3 Purified amplified library concentrations for positive and negative controls from both the manual and automated procedures were found to be within the acceptable limits for sequencing analysis, demonstrating the VERSA’s suitability for library preparations (Table 2). The SNP frequency of nucleotides determined from manual and automated performance was comparable, showing high correlation values across all libraries (Table 3). Manual and automated library correlations (r2) for both the positive and negative controls (Table 4) demonstrate the system’s high reproducibility; this can be attributed to the uniform distribution of magnetic beads and accurate pipetting.
Table 2 – Purified amplified library concentrations for positive and negative controls from the manual and automated procedures within the acceptable limits
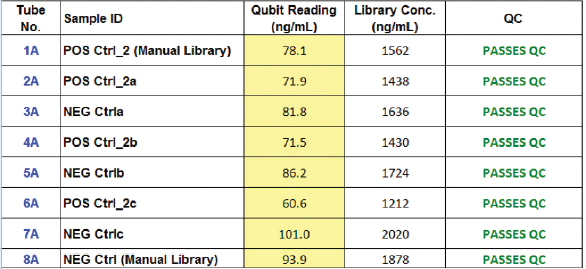
Table 3 – SNP frequency of nucleotides determined from manual and automated performance
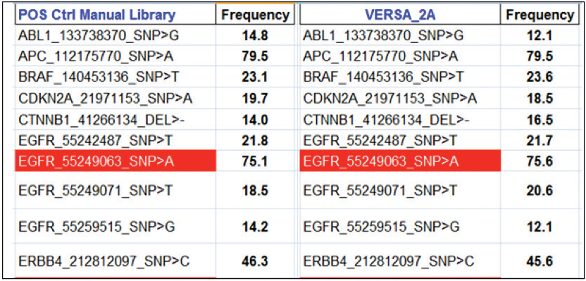
Table 4 – Correlation of manual and automated libraries
References
- Grada, A. and Weinbrecht, K. Next-generation sequencing: methodology and application. J. Investigative Dermatology 2013, 133, 248–56.
- Schneeberger, K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nature Rev. Genetics 2014, 15, 662–76.
- Zhang, L.; Chen, L. et al. Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing. Oncologist 2014, 19(4), 336–43.
- van Wezel, E.M.; Zwijnenburg, D. et al. Whole-genome sequencing identifies patient-specific DNA minimal residual disease markers in neuroblastoma. J. Molec. Diagnostics 2015, 17(1), 43–52.
- Gill, S.; Gill, R. et al. Automated bead mixer for VERSA Mini Nucleic Acid Purification/PCR setup workstation. Laboratory Focus July 2011, 8–10.
- Rodriguez, R. and Miller, K.M. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nature Rev. Genetics 2014, 15, 783–96.
- Zentner, G.E. and Henikoff, S. High-resolution digital profiling of the epigenome. Nature Rev. Genetics 2014, 15, 814–27.
- Schlötterer, C.; Tobler, R. et al. Sequencing pools of individuals—mining genome-wide polymorphism data without big funding. Nature Rev. Genetics 2014, 15, 749–63.
- Schneeberger. K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nature Rev. Genetics 2014, 15, 662–76.
- Lasken, R.S. and McLean, J.S. Recent advances in genomic DNA sequencing of microbial species from single cells. Nature Rev. Genetics 2014, 15, 577–84.
- Veeramah, K.R. and Hammer, M.F. The impact of whole-genome sequencing on the reconstruction of human population history. Nature Rev. Genetics 2014, 15, 149–62.
- Rogers, J. and Gibbs, R.A. Comparative primate genomics: emerging patterns of genome content and dynamics. Nature Rev. Genetics 2014, 15, 347–59.
- Ingolia, N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nature Rev. Genetics 2014, 15, 205–13.
- Fricke, W.F. and Rasko, D.A. Bacterial genome sequencing in the clinic: bioinformatic challenges and solutions. Nature Rev. Genetics 2014, 15, 49–55.
- Shapiro, E.; Biezuner, T. et al. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Rev. Genetics 2013, 14, 618–30.
- Mwenifumbo, J.C. and Marra, M.A. Cancer genome-sequencing study design. Nature Rev. Genetics 2013, 14, 321–32.
- Rehm, H.L. Disease-targeted sequencing: a cornerstone in the clinic. Nature Rev. Genetics 2013, 14, 295–300.
- Veltman, J.A. and Brunner, H.G. De novo mutations in human genetic disease. Nature Rev. Genetics 2012, 13, 565–75.
- Cho, I. and Blaser, M.J. The human microbiome: at the interface of health and disease. Nature Rev. Genetics March 2012, 13(13), 260–70.
- König, J.; Zarnack, K. et al. Protein–RNA interactions: new genomic technologies and perspectives. Nature Rev. Genetics 2011, 12, 745–55.
- Bamshad, M.J.; Ng, S.B. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature Rev. Genetics 2011, 12, 87–98.
- Ozsolak, F. and Milos, P.M. RNA sequencing: advances, challenges and opportunities. Nature Rev. Genetics 2011, 12, 7–18.
- Zhou, V.W.; Goren, A. et al. Charting histone modifications and the functional organization of mammalian genome. Nature Rev. Genetics 2010, 11, 9–16.
- Sorek, R. and Cossart, P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nature Rev. Genetics 2010, 11, 9–16.
The authors are with Aurora Biomed Inc., 1001 E. Pender St., Vancouver, B.C., Canada V6A 1W2; tel.: 604-215-8700; e-mail: [email protected]; www.aurorabiomed.com. The authors acknowledge the support of the Virginia Commonwealth University Molecular Pathology Laboratory (Richmond, Va.) for aiding in the sequencing experiments and assisting in the NGS sequencing and analysis.