The traditional “scales of justice” represent truth and fairness. For the scientific community, the balance is between big data (terabytes of data across numerous databases) on one scale and reproducible science on the other. The wealth of information coming out of ’omics-based (genomics, proteomics, cellomics, metabolomics, etc.) research in combination with patient-related outcomes has grown exponentially. Increasingly, the scientific community is dependent on the verifiability of the science that is generated with these data. Assumptions are being made that the published conclusions are accurate and will stand the test of time. However, this often is not the case.1
Traditional methods for ensuring data reproducibility are coming under increased scrutiny, particularly with the pressures of translating basic research into clinical outcomes and therapeutics.2,3 While technical replicates and positive and negative controls are built into the experimental design to mitigate experimental variability and enhance the clarity of the results, the question should be asked: Is this the most efficient way to increase the robustness of the data and advance the pace of basic research or translational science?
Given the complexity of the molecular mechanisms of disease and therapeutics, no research lab can single-handedly generate the breadth of data necessary to advance translational science (using the basic research building blocks to further our understanding of biological processes and develop new diagnostic tools, therapies, or medical procedures). As a result, consortia of different kinds—inter- and intra-institutional collaborations in academia and between academia and industry—have become the norm. In addition, huge amounts of information are being imported daily into global databases from a variety of research labs; data are being shared and redistributed instantaneously. However, this puts increased pressure on the need for “verifiable science” and being able to trust the accuracy and validity of the data upon which subsequent experiments are based.4 This pressure necessitates a standardized means for comparing results within and across labs.2
Achieving method standardization in the translational and research lab
When preparing tens and hundreds of biological samples for analysis, a focus on validated reagents, standardized methodologies, and calibrated equipment is critical. Method consistency within and between labs can be made more reproducible when routine pipetting tasks are separated from user variability. Method consistency, without the need for additional lab personnel, can be achieved with PIPETMAX®, a small, automated “lab assistant” pipetting station (Gilson Inc., Middleton, WI). Surpassing the consistency of even experienced pipet operators, PIPETMAX (Figure 1) can provide the method standardization and reproducible results required for basic research and translational labs.
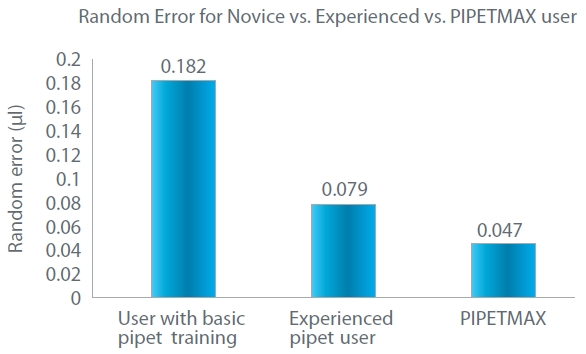 Figure 1 – Comparison of random pipetting error for novice pipet user vs experienced PIPETMAN user vs PIPETMAX.
Figure 1 – Comparison of random pipetting error for novice pipet user vs experienced PIPETMAN user vs PIPETMAX.By preparing every assay with proven consistency, PIPETMAX delivers high precision and accuracy in routine pipetting tasks, minimizes cross-contamination of samples, and maximizes reproducibility of the data and productivity within a lab and across collaborative sites. Every PIPETMAX comes with the precision of the PIPETMAN® (Gilson) inside pipetting head, which delivers reliable and consistent pipetting time after time from plate to plate. With the reduced need for technical replicates to compensate for pipetting inconsistencies and other experimental variability, the wastage in samples, time, and reagents is minimized with PIPETMAX.
Minimizing the deleterious impact of contaminants from the lab environment is equally important to data reproducibility; PIPETMAX can be used inside a sterile hood, where an external sensor detects when the hood is closed. Alternatively, the standalone benchtop system has an accompanying enclosure to avoid lab contaminants. For performance assurance, on-instrument calibration of the pipetting heads and alignment of the XYZ platform are achieved with the respective calibration and alignment protocols.
Application for quantitative polymerase chain reaction (qPCR) and PCR
With the increasing need for biomarker validation using qPCR and PCR techniques across a variety of biological samples, researchers in translational science are seeking new ways to maximize qPCR sample preparation accuracy and reproducibility, eliminate inherent variability, and minimize sources of contamination. PIPETMAX qPCR Assistant software is a workflow-based solution used with the PIPETMAX for creating and running automated PCR and qPCR experiments. With a versatile qPCR-focused design of experiment workflow, the researcher focuses on creating sophisticated or simple qPCR protocols by answering simple questions about the qPCR experimental design instead of addressing the typical liquid handling automation details. Performing routine and standardized qPCR experiments is simplified with drop-down menus and predesigned methods.
PIPETMAX performs every action reproducibly, eliminating variability in touch-off, purge, and pipet stroke movements that could lead to inconsistent liquid handling. Each step of the qPCR protocol—sample dilution, master mix preparation, sample transfers, and master mix transfers—is optimized based on its unique liquid handling needs. This means that additional liquid handling optimization, previously one of the most difficult and time-consuming steps in method automation, is no longer necessary.
The efficiency and traceability of qPCR preparations are improved because qPCR Assistant:
- Imports sample identity information and exports reaction plate templates to save time and ensure error-free traceability
- Creates single- or multiplex qPCR protocols to provide effortless flexibility
- Combines multiple qPCR methods into a single run to increase efficiency.
With the ability to guide the researcher on the labware positions and how and what to prepare for each run, the software then exports a template file for the most common thermocyclers. The optimized qPCR plate setup function enables method and pipetting efficiency, decreasing run times and increasing method traceability, thus providing the reassurance of reproducible results.
To ensure best practices are consistently incorporated into research methods and monitored electronically, even for labs that do not have electronic notebooks or LIMS, the qPCR Assistant software generates a traceable output report that provides the experimental design details and plate locations for the thermocycler. In addition, the traceability provides an added level of electronic security, unavailable when experiments are done manually, by helping to eliminate manual errors, maximizing data reproducibility, and further saving the researcher time, thereby strengthening the ability to troubleshoot and provide a standardized means for comparing results within and across labs.
Conclusion: Advancing the pace of basic research and translational science
Ultimately, the goal is to increase the pace of scientific discovery by trusting the existing scientific data upon which the future is being built. Method standardization and data traceability using applications-based automated “lab assistants” and software are necessary to keep the scales of scientific justice in check. Enabling scientists in academia and industry to maintain higher standards of data reproducibility will result in increased productivity and help to lower the barriers to translational science and improve patient outcomes.
References
- Begley, C.G.; Ellis, L.M. Drug development: raise standards for preclinical cancer research. Nature 29 Mar 2012, 483, 531–3.
- The Case for Standards in Life Science Research: Seizing Opportunities at a Time of Critical Need; 2013 survey report, Global Biological Standards Institute.
- Global Survey of Academic Translational Researchers: Current Practices, Barriers to Progress, and Future Directions; 2013 survey report, AAAS–Sigma Aldrich.
- Wadman, M. NIH mulls rules for validating key results: US biomedical agency could enlist independent labs for verification. Nature 1 Aug 2013, 500, 14–16.
Tristan Berto is the Product Manager for automated liquid handling and SPE instrumentation at Gilson, Inc., 3000 Parmenter St., Middleton, WI 53562, U.S.A.; tel.: 608-828-3278; e-mail: [email protected]; www.gilson.com