From the discovery of new miRNAs to clarifying their biological roles and utility as biomarkers, micro RNA (miRNA)-based research is undergoing a period of change. Rigorous methods are therefore required to profile large sample cohorts.
SmartRNAplex™ assays (EMD Millipore, Temecula, CA) address this need by providing a method for multiplexed miRNA analysis of up to 68 targets across as many as 96 samples. With SmartRNAplex customizable assays, researchers are able to design panels that detect any combination of miRNAs annotated in miRbase (from any species). The assays have been used to profile miRNAs in human, mouse, rat, and C. elegans over a broad range of sample types, including serum, plasma, crude cell digests, and formalin-fixed tissues.
 Figure 1 – SmartRNAplex workflow. The SmartRNAplex assay is carried out in standard 96-well filter plates. After miRNA targets are hybridized to hydrogel particles and labeled, fluorescent reporter is added and assay readout is performed using a standard flow cytometer. Data files from the cytometers are interpreted in Firefly software for analysis and export.
Figure 1 – SmartRNAplex workflow. The SmartRNAplex assay is carried out in standard 96-well filter plates. After miRNA targets are hybridized to hydrogel particles and labeled, fluorescent reporter is added and assay readout is performed using a standard flow cytometer. Data files from the cytometers are interpreted in Firefly software for analysis and export.Multiplexed assay platform
The SmartRNAplex platform is a multiplexed assay that is able to measure several targets simultaneously in a single sample. Multiplexing is accomplished by using encoded hydrogel Firefly™ particles (Firefly BioWorks, Cambridge, MA). Each particle bears a unique “bar code” that identifies the miRNA species for which that particle bears a complementary binding sequence. The particles are manufactured via optical liquid stamping—a method for microparticle fabrication that enables the synthesis of microscopic structures of virtually any shape and chemical functionality.
Including hands-on time, the assay requires 3.5–4.5 hours from assay setup to data, depending on how many samples are processed in parallel (Figure 1). Unlike other systems that are dependent on glass or polystyrene substrates, the particles are composed of bio-inert poly (ethylene glycol) hydrogels. The substrate provides solution-like thermodynamics for optimal sensitivity and specificity, and enhanced capacity for three-dimensional target capture, leading to a greater dynamic range. Coupled with the SmartRNAplex posthybridization labeling method, the substrate makes the platform well suited for applications that require detection in crude samples.
Posthybridization labeling
Traditionally, in order to profile miRNAs in complex samples such as cell lysate, plasma, and serum, RNA purification is necessary.This often requires laborious protocols and the use of toxic chemicals, and may lead to biases in target recovery. In order to overcome this limitation, a posthybridization labeling method was developed for the detection of miRNA targets directly from crude samples, regardless of purity (Figure 2). In addition, bound targets can be labeled on either the 3’ or 5’ end to discriminate mature miRNAs from precursor species.
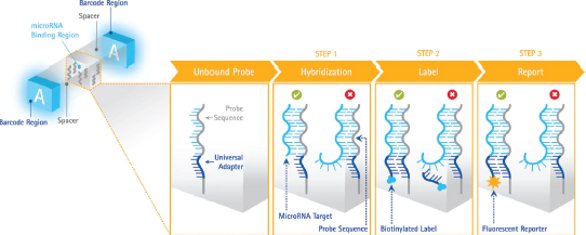 Figure 2 – Posthybridization labeling scheme. Left: Probes embedded throughout the hydrogel particle have sites for a specific miRNA and an adjacent adapter site for a universal label, which is detected via fluorescence. Right: Assay workflow involves hybridization for target capture, attachment of a universal adapter via labeling, and reporting with fluorescence. Target sequence binding (green check mark) results in a signal, while nonspecific RNAs or precursor miRNAs (red X) do not generate a signal.
Figure 2 – Posthybridization labeling scheme. Left: Probes embedded throughout the hydrogel particle have sites for a specific miRNA and an adjacent adapter site for a universal label, which is detected via fluorescence. Right: Assay workflow involves hybridization for target capture, attachment of a universal adapter via labeling, and reporting with fluorescence. Target sequence binding (green check mark) results in a signal, while nonspecific RNAs or precursor miRNAs (red X) do not generate a signal.The SmartRNAplex assay labels targets after they have been captured by miRNA-specific probes embedded in the hydrogel particles. Probes have two binding sites: one that binds a specific miRNA, and the other that binds a universal adapter sequence used for labeling (Figure 2, left). When an miRNA target is captured on its corresponding probe, a universal adapter is attached to that miRNA via ligation. After addition of a reporter species, this binding event is detected via fluorescence. The level of fluorescence is quantitative, providing an accurate indication of target levels in a given sample.
Assay workflow
The assay is performed in a 96-well filter plate that permits the simultaneous analysis of up to 96 samples. All assay steps are carried out at 37 °C or at room temperature with shaking. All reagents are nontoxic, yet provide stringent hybridization conditions for highly specific miRNA detection. The basic assay workflow involves five main steps with intermittent rinsing:
- Mix: addition of particles, hybridization buffer, and samples to assay wells
- Hybridize: 90-min incubation at 37° C; 2× rinse
- Label: addition of labeling buffer, followed by 45-min incubation at room temperature; 2× rinse
- Report: addition of reporter, followed by 45-min incubation at room temperature; 2× rinse
- Scan: addition of run buffer, followed by scanning on a standard flow cytometer.
Flow cytometer assay readout
The final assay readout can be performed with a standard flow cytometer. The minimum configuration of the system is a single blue laser (~488 nm) for excitation with green (~525 nm), yellow (~580 nm), and red (~690 nm) detectors.
 Figure 3 – Data interpretation using Firefly software. Shown on the left are scatter plots obtained by scanning Firefly particles on the guava easyCyte™ 8HT cytometer (EMD Millipore). The multiple events captured for each particle are interpreted using Firefly software to identify codes and quantify targets (right)
Figure 3 – Data interpretation using Firefly software. Shown on the left are scatter plots obtained by scanning Firefly particles on the guava easyCyte™ 8HT cytometer (EMD Millipore). The multiple events captured for each particle are interpreted using Firefly software to identify codes and quantify targets (right).
Firefly particles are designed to appear to the cytometers as a series of closely spaced cells, and each particle is recorded as multiple sequential events (Figure 3). The high aspect ratio of the particles is optimized in order to force alignment with flow in a cytometer flow cell, even in the absence of sheath flow. Using the information contained within the multiple regions of the particles, as many as 70 codes (68 targets plus 2 controls) can be distinguished reliably. For specialty applications, the particles can be designed to work with much simpler devices that have only a single excitation source and detector.
For interpretation of the data collected on a standard flow cytometer, the Firefly software suite is used to analyze the data generated with the SmartRNAplex assay. Flow cytometry standard (FCS) files saved from the cytometer are uploaded into the software, along with a “plex” (“PLX”) file that contains information on the targets being detected for a given assay. The software parses through the events contained within the FCS file(s) and regroups them into particle information, with bar code data and target levels, in seconds. The data are presented in plots showing the heat map (Figure 3, top right) and target quantification (Figure 3, bottom right) for a given sample. Data can be exported for further analysis using other software.
miRNA profiling in cell lysates
Expression profiling of three breast cancer cell lines was performed. The results obtained using purified RNA were compared with those attained using crude cell lysate as the input material. For purified RNA samples, TRIzol® reagent (Life Technologies, Carlsbad, CA) was used with the recommended protocol. Because samples were abundant, approximately 2 μg total RNA was used in the assay. For lysate samples, cells were incubated for 60 min at 55 °C in Digest Buffer, after which samples were filtered and 25 μL was added directly into the assay. The cell equivalent for lysate experiments was approximately 10,000 cells.
 Figure 4 – Equivalent miRNA profiling from purified RNA and cell lysates. Heat maps show log2 transformed, miR-16-normalized data (left). Gray cells represent targets that were not detected. Shown on the right is a scatter plot comparing miR-16 normalized data across the two data sets.
Figure 4 – Equivalent miRNA profiling from purified RNA and cell lysates. Heat maps show log2 transformed, miR-16-normalized data (left). Gray cells represent targets that were not detected. Shown on the right is a scatter plot comparing miR-16 normalized data across the two data sets.After analysis, data were normalized to miR-16 levels for comparison. As shown in Figure 4, the profiling data for purified RNA closely matched those for the crude cell digests. This simple protocol enables analysis of cells without the need for toxic chemicals or laborious phase separations.
Conclusion
The SmartRNAplex assay provides a means with which to profile miRNA in any species across a broad range of starting material. The multiplexed assay is customizable for the detection of up to 68 miRNA targets per sample with a rapid workflow. Researchers can easily discover which miRNA targets are relevant without the limitations of traditional methods. The assay also demonstrates consistent performance and, in all cases, detection limits were near or lower than 1 attomole and the assay showed linearity over 5 logs of dynamic range.
Additional reading
- Chapin, S.C.; Pregibon, D.C. et al. Rapid miRNA profiling on encoded gel microparticles. Angew. Chem. Int. Ed. 2011, 50, 2289–93.
- Chapin, S.; Pregibon, D.C. et al. High-throughput flow alignment of barcoded hydrogel microparticles. Lab Chip 2009, 9, 3100–9.
- Pregibon, D.C.; Doyle, P.S. Optimization of encoded hydrogel particles for nucleic acid quantification. Anal. Chem. 2009, 81, 4873–81.
- Pregibon, D.C.; Toner, M. et al. Multifunctional encoded particles for high-throughput biomolecule analysis. Science 2007, 315, 1393–6.
- Dendukuri, D.; Pregibon, D.C. et al. Continuous flow lithography for high-throughput microparticle synthesis. Nat. Mater. 2006, 5, 365–9.
Mark Santos is R&D Manager, EMD Millipore, 28820 Single Oak Dr., Temecula, CA 92590, U.S.A.; tel.: 951-514-4538; email: mark.santos@ emdmillipore.com ; www.emdmillipore.com