Precise and biologically relevant in vitro studies require the ability to analyze cell responses to perturbations over time.1 In order to achieve successful long-term cell culture and imaging, however, technical challenges must be overcome. It is difficult to control the temperature and gas composition of the cell environment without obstructing optical access to the cells, and it is also challenging to sustain cell health by maintaining sufficient outflow of waste and nutrient supply.
The CellASIC™ ONIX Microfluidic Platform from EMD Millipore (Boston, MA) offers a solution by delivering precise microenvironmental control of perfusion, gas, and temperature conditions. This facilitates long-term cell culture and high-quality, live cell microscopy, enabling automated, in-plate image analysis with minimal reagent volumes. The microfluidic chamber recreates the physiologic mass transport condition for optimized cell health, and an integrated microincubator system delivers gas and temperature control. Four upstream fluidic channels allow controlled exposure of the cells to different solutions during live imaging. With a dedicated gravity-driven flow channel, the plate can also be cultured in a standard incubator. The cells contact an optical glass surface, enabling high-quality imaging using an inverted microscope.
 Figure 1 – Equal, even pressure-driven cell loading. All four cell chambers of the M04S plate were primed with 0.1% gelatin solution, via capillary action, prior to cell loading. Suspensions of MCF7 cells were prepared at 1.0 × 106 cells/mL (for chambers A and B) or at 2.0 × 106 cells/ mL (for chambers C and D). Inlet well 6 was set to flow at 0.25 psi for 0.1 min (step 1 of the entire protocol as shown in Figure 2) for pressure-driven cell loading.
Figure 1 – Equal, even pressure-driven cell loading. All four cell chambers of the M04S plate were primed with 0.1% gelatin solution, via capillary action, prior to cell loading. Suspensions of MCF7 cells were prepared at 1.0 × 106 cells/mL (for chambers A and B) or at 2.0 × 106 cells/ mL (for chambers C and D). Inlet well 6 was set to flow at 0.25 psi for 0.1 min (step 1 of the entire protocol as shown in Figure 2) for pressure-driven cell loading.This article demonstrates the long-term culture of multiple cell lines using automated perfusion protocols set up with CellASIC ONIX FG software, and compares the results to those obtained using traditional chambered slides. The results indicate that optimal experimental parameters—such as flow rates, flow duration, extracellular matrix (ECM) concentration, gas composition, and medium change intervals—can vary greatly with respect to the particular cells being cultured.
The article also demonstrates live/dead staining of cells and automated immunostaining of cells within the microfluidic plate, showing that these automated protocols save time and can conserve antibodies and staining reagents.
Materials and methods
The CellASIC ONIX Microfluidic Platform was used for the long-term perfusion of three cell lines: NIH 3T3, MCF7, and PC-3. The CellASIC ONIX Microfluidic System delivered a controlled perfusion of media to the cells, while the CellASIC ONIX Microincubator Controller provided the microenvironmental conditions required for healthy cell culture. The CellASIC ONIX M04S Microfluidic Plate and manifold with recirculating heater were kept on the microscope stage to allow for live cell imaging of the cell culture. Cell proliferation was monitored and compared to a control culture. The three cell lines were cultured, and the cells were then trypsinized, harvested by centrifugation, and resuspended in complete medium.
Cell loading
All four cell chambers of an M04S plate were primed with media containing 10% fetal bovine serum (FBS) (for NIH 3T3 cells) or 0.1% gelatin (for MCF7 and PC-3 cells) via capillary action. The temperature calibration plate was sealed to the CellASIC ONIX manifold and calibrated to ensure the experiment would occur at the desired temperature. Cell suspension was added to each cell inlet.
Programming automated perfusion culture
The CellASIC ONIX FG software was used to create automated protocols (Table 1) that maintained healthy long-term cultures of NIH 3T3, MCF7, and PC-3 cells in the M04S plate. The software allows boxes to be checked to define the flow source, flow rate, and flow duration for each step of the protocol.
Cells and media were preloaded into the cell inlet and solution inlet wells, respectively, of the M04S plate. An uninterrupted protocol was created in which inlet well 6 was set to flow at 0.25 psi for 0.1 min, for pressure-driven loading of cells. To achieve continuous perfusion, each solution inlet well (wells 2–5) was filled with 300 μL medium, allowing scheduled medium flow from different solution inlets throughout the experiment. Flow rates for inlet wells 2–5 were set to 1 psi for NIH 3T3 cells and 0.25 psi for MCF7 and PC-3 cells. The M04S plate was sealed to the manifold, which connected the plate to the microfluidic system and microincubator controller. The recirculating heater was set to 37 °C. The plate was placed on the microscope stage to visualize cell loading and to monitor long-term cell culture outside the incubator.
Table 1 – Summary of perfusion protocols for cell lines used
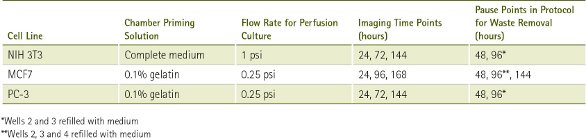
Images were acquired during perfusion at periodic intervals. Also, the program was paused at two or three points and excess medium was removed from the outlet wells. If needed, the medium in solution inlet wells was replenished before resealing the M04S plate to the manifold.
Live/dead staining of MCF7 cells within the microfluidic plate
The LIVE/DEAD® Viability Kit (Life Technologies, Grand Island, NY) was used, in which live cells fluoresce green and dead cells fluoresce red. The M04S microfluidic plate was unsealed from the manifold and the temperature was set to room temperature. The wells to be used for live/dead staining were aspirated. Live/dead stain was added to wells A2, C2, A4, and C4. Phosphate-buffered saline (PBS) was added to wells A3, C3, A5, and C5. The plate was sealed to the manifold and the software was used to create and run the automated staining protocol. A two-step protocol was created in order to flow live/dead stain, followed by PBS, into the cell chambers:
Step 1 (live/dead stain): Valves V2 and V4, pressure = 0.5 psi, 60 min
Step 2 (PBS): Valves V3 and V5, pressure = 0.5 psi, 5 min.
Automated immunostaining of PC-3 cells
The manifold was set to room temperature and immunostaining solutions were prepared. Inlet wells used for immunostaining were aspirated and filled. The plate was sealed to the manifold and the software was used to create and run the automated staining protocol.
Traditional cell culture
All three cell lines were also cultured using a Millicell® EZ slide (EMD Millipore). After 168 hr, these MCF7 cells also underwent viability staining using the LIVE/DEAD stain.
Microscopy
All images were acquired with a Q Color 5 Camera (Olympus, Center Valley, PA) attached to an Axiovert® inverted microscope (Carl Zeiss, Peabody, MA), using ImagePro software (Image-Pro Software, Rockville, MD). ImageJ software (Image-Pro Software) was used to merge images.
Results
Equal, even pressure-driven cell loading into chambers of microfluidic plates
It is only possible to make biologically relevant conclusions by comparing one cell culture chamber to another in a single multichamber plate if cells are evenly loaded and subject to the same perfusion conditions. NIH 3T3, MCF7, and PC-3 cells were loaded into replicate chambers of the microfluidic plate. The resulting even distribution of the cells, not only across each chamber surface, but also when comparing each chamber to the next, illustrated the precision with which the flow from the inlet wells of the microfluidic system can be controlled. Figure 1 shows MCF7 cells for representative results.
 Figure 2 – Long-term cell culture of MCF7 cells in the CellASIC ONIX Microfluidic Platform (left) compared to static culture in the Millicell EZ slide (right). Even after 168 hr of culture, cells looked qualitatively healthy and showed similar proliferation rates under both culture conditions.
Figure 2 – Long-term cell culture of MCF7 cells in the CellASIC ONIX Microfluidic Platform (left) compared to static culture in the Millicell EZ slide (right). Even after 168 hr of culture, cells looked qualitatively healthy and showed similar proliferation rates under both culture conditions. Figure 3 – Live/dead staining of long-term cultures of MCF7 cells in the CellASIC ONIX Microfluidic Platform (left) compared to static culture in the Millicell EZ slide (right). Even after 168 hr of culture, cells showed high viability under both culture conditions.
Figure 3 – Live/dead staining of long-term cultures of MCF7 cells in the CellASIC ONIX Microfluidic Platform (left) compared to static culture in the Millicell EZ slide (right). Even after 168 hr of culture, cells showed high viability under both culture conditions.Comparison of automated perfusion culture to traditional, static culture
The long-term perfusion cultures of NIH 3T3, MCF7, and PC-3 cells were compared with static, incubator cultures of these same cells on traditional chambered slides. For all three cell lines, the cells showed similar morphology and qualitative proliferation rate for both conditions (MCF7 cells shown as representative line, Figure 2). Live/dead staining of MCF7 cells showed very good viability on both culture platforms after 168 hr of culture (Figure 3).
Facilitating the immunocytochemistry (ICC) work flow
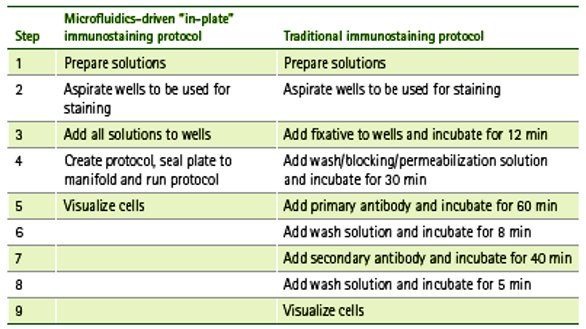
An automated immunostaining protocol was created that decreased the hands-on time traditionally required for immunostaining. As shown in Table 2, in-plate immunostaining required 44% fewer hands-on steps than traditional immunostaining.
Table 2 – Fewer hands-on steps required for in-plate, microfluidics-driven immunocytochemistry (left column) compared to traditional immunocytochemistry (right column)
Using automated protocols and perfusion culture as well as manual staining and static culture, the ICC of PC-3 cells showed robust CD9 and cytokeratin 19 expression and visible nuclei. Qualitatively, the staining patterns are similar; however, the spatial dimensions of the microfluidic cell culture chamber places more cells within the focal plane of the microscope, enabling higher-resolution images of more cells.
Conclusion
These analyses have shown that it is possible to maintain a virtually hands-free, healthy cell culture for many days using perfusion feeding. The cell cultures in microfluidic plates were healthy, proliferated well, and exhibited similar morphology and viability to parallel cultures kept under static conditions in a traditional incubator.
The perfusion conditions presented here are optimized, obtained by testing different cell lines with varying flow rates, durations, etc. The authors recommend that users optimize these conditions for their own cells and experimental perturbations. Their specific observations include:
- Pressures between 0.25 and 1 psi were ideal, corresponding to lower flow rates and providing adequate nourishment with minimal mechanical stress
- Some cell types needed ECM coating to facilitate cell adhesion
- Optimal initial cell concentration depends on cell types and duration of culture. Here, concentrations of 1 × 106 and 2 × 106 cells/mL were used for long-term cell culture
- Loading conditions may need optimization, depending on cell type and cell concentration used
- During long-term culture, waste cells need to be emptied periodically and solution wells replenished. Because the automated perfusion program can be paused and resumed, these steps are easily done.
The CellASIC ONIX platform also facilitates automated immunostaining for both extracellular and intracellular proteins. The same concentrations of antibodies were used for the microfluidic plate and chambered slide, and similar staining patterns were observed in both analyses.
Reference
1. Sung, MH; McNally, J.G. Live cell imaging and systems biology. Wiley Interdiscip. Rev. Syst. Biol. Med.2011 Mar-Apr, 3(2), 167–82.
Kathleen Ongena, Ph.D., is R&D Manager; Sónia Gil is Biochemical Scientist; Cindy Chen is R&D Scientist; Paul Hung is R&D Manager; and Alex Mok is Product Manager, EMD Millipore, 17 Cherry Hill Dr., Boston, MA 01923, U.S.A.; tel.: 978-762-5324; e-mail: [email protected]; www. emdmillipore.com.