Key protein complexes are fundamental to a wide range of cellular processes and key metabolic pathways within the body. Their dysfunction is associated with a range of disease pathologies such as cystic fibrosis, Alzheimer’s disease, and Parkinson’s disease.1,2 The study of the molecular structure and function of multiprotein complexes is therefore of value for the successful determination of their role in biological pathways and for the development of diagnostics and therapeutics. However, for this to be possible, techniques are needed that can increase both the yield of protein and the number of complexes that can be produced.
This article describes an automated method for optimizing expression of multiprotein complexes in insect cells using the advanced microscale bioreactor (ambr™) system (TAP Biosystems, Royston, U.K.). The application of laboratory automation can facilitate medium- to high-throughput multicomplex protein expression programs and, when combined with good experimental design, shows that protein expression can be rapidly improved compared to the traditional shake flask method.
Study of eukaryotic multiple protein complexes
For successful molecular and structural studies of protein complexes, it is necessary to obtain large quantities of protein. However, the low abundance of these endogenous protein complexes, as well as their inherent instability following successful isolation, commonly inhibits molecular and structural studies. Advances in molecular biology and the use of recombinant biology techniques over the past decade for the overexpression of such proteins have been invaluable. Following the identification and isolation of the genetic material encoding these protein complexes, there are a host of prokaryotic and eukaryotic systems available for their expression and synthesis.3
Currently the most widely employed systems for the isolation and study of recombinant human multiprotein complexes are baculovirus expression vector systems (BEVS), which employ insect cell lines as the host cell for protein expression. This system provides the eukaryotic host machinery necessary to support correct post-translational modification for authentic processing, with correct structural conformation and activity of the protein complex. With the ability to insert multiple genes into the baculovirus vector, infected insect cell lines can be established that express multiple protein subunits and hence multiprotein complexes. BEVS also enable in situ engineering of individual protein components, making it possible to express a wide range of protein complex variants.
In order to establish, grow, and maintain an insect cell culture in suspension, specific oxygen, osmolality, nutrient, pH, and temperature are required. Continual monitoring of growth conditions is necessary to ensure successful infection and recombinant protein overexpression. Insect cell line cultures require the temperature to be maintained within 25–28 oC, and it is essential that insect cells are in log phase at the time of infection. In order to ensure cells are adequately oxygenated and remain in suspension, traditional methods that use shake or spinner flasks are commonly employed.
Issues with recombinant expression systems for multiprotein complexes
BEVS such as the MultiBac, developed by the Berger laboratory at the European Molecular Biology Laboratory (EMBL, Grenoble, France), are widely used for the expression of large eukaryotic complexes with many genes encoding subunits.4 The MultiBac system enables in situ gene manipulation and the study of multiple variants for screening purposes. Not only can the simultaneous establishment, growth, and study of large numbers of protein variants following infection with multiple viral clones prove costly and time consuming, but also pose difficulties for manual handling and monitoring.
Culturing dozens of protein variants in parallel demands adequate space in order to house large shaker platforms. The use of traditional shake flasks for such studies requires considerable reagent volumes and manual labor for experimental setup, continual monitoring of growth conditions, and subsequent protein expression studies. Furthermore, as with all experiments involving large sample numbers, manual error is inevitable. In summary, the use of shake flask systems does not readily provide the flexibility for the simultaneous study of a large number of altered recombinant multiprotein complexes and cell line variants in a screening program.
Applications of the bioreactor system
Monitoring of cell cultures
 Figure 1 – ambr automated micro bioreactor system.
Figure 1 – ambr automated micro bioreactor system.The automated ambr micro bioreactor system overcomes the drawbacks surrounding the simultaneous culturing of large numbers of cell line variants. With up to 24 or 48 miniaturized bioreactors containing 10–15 mL of media (Figure 1), ambr enables the successful small-scale production of a large number of cultures, with less effort and potential for error. With active cooling, the culture can be set and maintained at a temperature of 27 oC, which is important for the successful infection and maintenance of insect cell lines.
The ambr workstation and flexible software enable automated liquid handling for media filling, inoculation, reagent or feed addition, cell sampling, splitting, and virus infection. Alongside monitoring and controlling culture conditions, the system eliminates the daily manual tasks of culture maintenance and sample tracking. With the ability to add nutrients, reagents, or cells without disturbing the continual movement of the impellers stirring the culture, minimal disruption of cell growth is achieved.
The workstation operates inside a biosafety cabinet, providing a reliable platform for sterile liquid handling such as feeding and sampling. Automated monitoring of dissolved oxygen and pH level through sensor spots found on the ambr bioreactors is also beneficial to cell growth. Active control of these two culture parameters inside each individual bioreactor provides the potential to optimize and enhance culture growth.
Bioreactor system software and footprint
In addition to providing continual culture monitoring, the system’s software can enable reports to be processed and transmitted to e-mail for remote monitoring at any time of the day or night. The ambr workstation allows control of feeding, sampling, and maintenance of environmental conditions for each individual bioreactor, providing the ability to study the effect of a range of conditions on a cell line or a range of recombinant virus clones. With up to 24 or 48 bioreactors per workstation, the system also provides the ability to include more control replicates for experimental and quality control.
With a small footprint—the size of a laboratory bench—the ambr workstation significantly reduces the amount of laboratory space required. With lower volumes, it also significantly reduces media and reagent costs.
Proof of concept of automation for high-throughput multiprotein expression
In a recent collaborative study carried out by TAP Biosystems and EMBL,5 two insect cell lines (Sf21 and Hi5) were cultured in triplicate in GIBCO (Life Technologies, Carlsbad, CA) and Hyclone Medium (Thermo Fisher Scientific, Waltham, MA) in the ambr system. The results confirm that the uninfected insect cell lines can be successfully cultured and maintained in an automated micro bioreactor system (Figures 2 and 3).
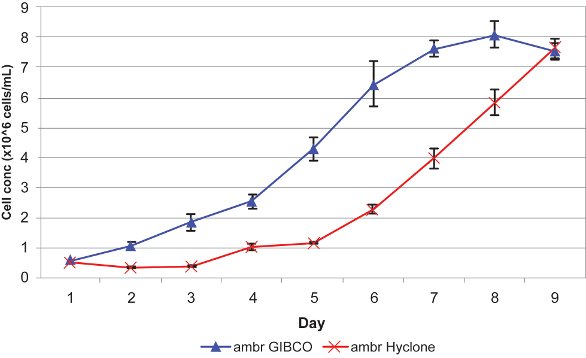
Figure 2 – Growth of Sf21 cells in ambr bioreactors cultured in triplicate GIBCO and Hyclone Medium.
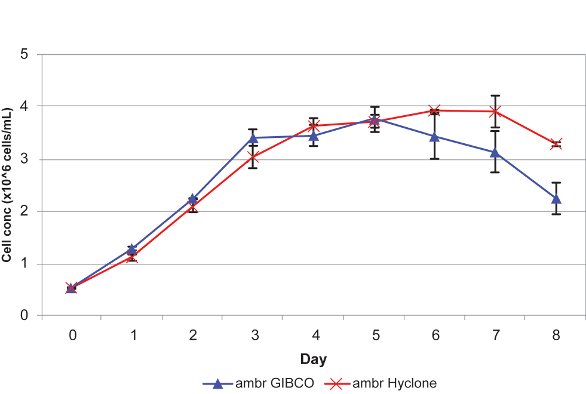
Figure 3 – Growth of Hi5 cells in ambr bioreactors cultured in triplicate GIBCO and Hyclone Medium.
A subsequent viral infection study of the cells using 12 ambr vessels and three shake flasks seeded with Sf21 cells and cultured for up to eight days was then carried out. Cultures were diluted 2:1 to achieve target cell density for virus infection. The ambr insect cell cultures were infected with a single baculovirus expressing a His-tagged 2-protein complex labeled with YFP (yellow fluorescent protein) and CFP (cyan fluorescent protein). Except for uninfected negative controls (condition A), the ambr system automatically seeded insect cell cultures, cell densities being 1.5 (D), 3.0 (C), or 6.0 (B) × 106 cells/mL, respectively, with the same virus such that the multiplicity of infection (MOI) remained the same in all bioreactors. The shake flask cell cultures were infected with a single baculovirus expressing a His-tagged 2-protein complex labeled with YFP and CFP at 1.5 (D, data unavailable), 3.0 (C), and 6.0 (B) × 106 cells/mL. Increasing YFP and CFP expression detected in both the manual shake flask and automated ambr culture systems over days 1–4 postinfection indicated successful viral infection (Figure 4).
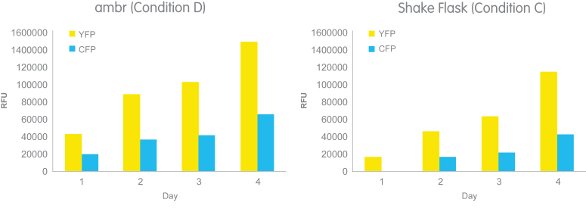
Figure 4 – CFP-YFP expression plots for ambr and shake flask cultures, four days postinfection starting with (D) 1.5 × 106 cells/mL for ambr and 3.0 × 106 cells/mL (C) for the shake flasks.
Following analysis of recombinant protein production in this study, the manual shake flask and automated ambr culture systems both indicated that YFP and CFP were produced in all the conditions tested (except the negative control), confirming successful infection (Figure 5). Indeed, the fact that the negative ambr control cultures (condition A) remained uninfected indicates that no viral cross-contamination occurred. Increased protein expression was observed from cultures grown in the ambr bioreactors infected at lower cell densities, and an infection cell density of 1.5 × 106 cells/mL was found to be near optimal. This is again confirmed by the shake flask cultures, with stronger expression observed from cultures infected at lower densities, matching earlier shake flask studies which suggested that infection between 0.5 and 2.0 × 106 cells/mL is optimal for insect cell protein expression.
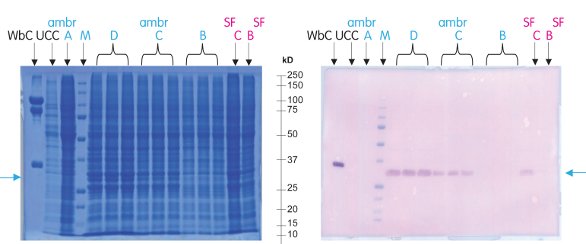
Figure 5 – SDS-PAGE and Western blot analysis of protein produced at CFY/YFP protein expression plateau (day 4). The target His-tagged 2-protein complex was positively identified against marker M (Precision Plus Protein™ All Blue Standards, Bio-Rad, Hercules, CA) and UCC (uninfected cell control).
The observed results confirm that sf21 insect cells can be successfully grown, maintained, and infected in the ambr bioreactors to express multiprotein complexes. The results also suggest that protein expression can be rapidly obtained with reduced effort, enabling speedy assessment of variants and culture parameters for high-throughput screening studies. With the ability to employ smaller culture volumes and monitor up to 24 or 48 bioreactors simultaneously, the automated system is well suited for the culture and study of multiple variants for rapid multiparallel protein expression.
High-throughput screening studies of multiprotein complexes
The study of large numbers of protein complex variants in parallel using manual shake flask methods is not only time consuming and expensive, but difficult to handle and evaluate. The automated workstation provides the ability to carry out high-throughput multiprotein expression screening studies, with minimal cost, time, and manual effort.
With the ability to simultaneously study up to 24 or 48 bioreactors per workstation employing low volumes of culture media per study, ambr enables multiple cell line selection and development experiments to be run simultaneously. This facilitates a wide range of parameter studies, such as the infectivity of viral clones, protein expression, and culture conditions in order to optimize insect cell protein expression and production. In addition, the system can be used to study the stability and productivity of expressed protein complexes across a range of clones in order to select the most promising variants before scaleup, or to study multiple replicates of the same clone to compare the effect of different feeding strategies or culture conditions on growth and expression levels.
In summary, this streamlining of protein expression studies using automated micro bioreactor systems will ultimately accelerate research on the structure and function of multiprotein complexes. The next step is to optimize the process to mimic larger-scale bioreactor cultures, as has successfully been achieved for mammalian cell cultures.6 Such studies are not only of interest for the understanding of the mechanisms and the role of these proteins in biological pathways, but also for the potential development of multiprotein diagnostics or protein therapeutics in future drug discovery programs.
References
- Guerriero, C.J.; Brodsky, J.L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 2012, 92(2), 537–76.
- Andre, R.; Tabrizi, S.J. Misfolded PrP and a novel mechanism of proteasome inhibition. Prion 2012, 6(1), 32–6.
- Rai, M.; Padh, H. Expression systems for production of heterologous proteins. Current Sci. 2001, 80, 1121–8.
- Berger, I.; Fitzgerald, D.J. et al. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004, 22(12), 1583–7.
- Chaillet M.; Garzoni, F. et al. MultiBac Expression System: comparison of growth and multiprotein production in shake flask and automated miniature bioreactor (ambr™) cultures. Poster presentation at ISBioTech 3rd Annual Meeting, Rosslyn, VA, Mar 11–15, 2013.
- Hsu, W.T.; Aulakh, R.P. et al. Advanced microscale bioreactor system: a representative scale-down model for bench-top bioreactors. Cytotechnology 2012, 64(6), 667–78.
Dr. Imre Berger is Group Leader, Structural Biology of Eukaryotic Complexes and Head of the Eukaryotic Expression Facility (EEF); Maxime Chaillet is Engineer for HT Technologies, EEF; and Frederic Garzoni is Facility Manager, EEF, European Molecular Biology Laboratory (EMBL), Grenoble Cedex 9, France. Dr. Sinyee Yau-Rose is a Bioprocess Product Specialist, and Dr. Barney Zoro is ambr Product Manager, TAP Biosystems, York Way, Royston, Herts SG8 5WY, U.K.; tel.: +44 (0) 1763 227200; e-mail: [email protected].