The biotechnology industry has been hit hard by the current financial crisis and faces a future where efficiency and collaboration will be prerequisites for survival. During the last eight months, one study reported that “120 companies announced restructuring plans, including reprioritizing their programs and reduction of workforce. . ..[Of] the 120, 100 are small biotech companies and 20 are major pharma or biotech companies.”1
“Do more with less” and “race against the clock” have become clichés in an industry already burdened with high risk as our brightest weave new threads into the fabric of scientific knowledge. The search is on for better tools that can automate discovery processes and yet provide the innovation needed to explore therapies for existing and emerging diseases. Additionally, especially in small biotech, productivity also means finding the right partner to take the torch of a novel discovery forward to commercialization, and ultimately a better quality of life for people treated with a new therapy. High-content technology, when used to provide more sophisticated cell-based assays, is one such productivity tool.
Cellomics Inc. (now part of Thermo Fisher Scientific, Pittsburgh, PA) introduced the first automated imaging platform for screening in the late 1990s, calling it High Content Screening (HCS). Over the last decade, the term “high content” has become synonymous with cell-based assays using multiparameter analysis of cell images as the detection method. High-content methods have become increasingly adopted across life science, and provide an unprecedented level of information about cellular responses. Taken together, these cell profiles are being used to improve target validation/identification, compound selection, ADME/tox, and even clinical candidate selection.
Automated imaging and analysis
Fluorescence microscopy has been around for a long time, but only relatively recently have advances in computing power, electronics, and robotics allowed a reproducible, flexible, and scaleable screening tool to be created. A number of features make this approach different from current plate reader assays, and imaging as a detection or quantitation method for cell-based assays provides a radically different way to assess compound effects.
Foremost is the ability to identify and measure individual objects in the well instead of just measuring the combined signal as a single output. Through automated analysis of the location and intensity of light signals from each image, cells, subcellular compartments, and multicellular structures can be identified using scientist-defined criteria. Importantly, any signal coming from objects outside the criteria is removed from analysis, enabling the rejection of artifacts and virtual isolation of cell subpopulations, both of which are impossible with standard plate reader assay formats.
The second major difference takes advantage of the core strength of fluorescence microscopy, i.e., the ability to measure multiple fluorescing probes in the same cell. To achieve this remarkable ability required a century of effort to develop both instrumentation and reagents optimized for discrete wavelengths across the visible spectrum. Automation of this ability to collect multiple signals per cell permits an enormous range of biologies to be turned into assays, and multiple phenotypes, signaling pathways, and toxicity indicators can often be simultaneously collected for each cell of a targeted population.
The third difference has much in common with success in real estate: location, location, location. Because each image delivers data as intensities at discrete locations (called pixels) a rich information “map” of each object emerges where size, shape, area, and texture can be quantified. These image features can be combined as assay outputs, or used as “masks” to define downstream locations to measure more targets. It is not uncommon for high-content assays to deliver hundreds of potentially useful measurements. Standard plate reader assays lose the cellular context when cell signals are homogenized, making these kinds of insights impossible.
The ability of high-content methods to automate unscaleable manual assays, create phenotype-based assessment, and allow novel targets to be addressed has emerged as an important factor in bridging the productivity gap in drug discovery. A significant number of major biotechnology companies have adopted this technology, and there is a growing compendium of peer-reviewed literature using high content in major research areas from basic cell signaling through therapy areas such as cancer, virology, and neurobiology to in vitro toxicology.
Cancer is a disease class where more commercially available, specific therapies are desperately needed. It is not surprising that high content is playing an important role in measuring the traditional targets (i.e., apoptosis, transformation, migration/metastasis, cell cycle), and also newer classes of targets such as autophagy,2 nuclear export,3 and kinase inhibition.4 High content can also be used to enhance the outcome of other cell-based technologies like RNA interference (RNAi).5
Viral infections not only directly account for significant sickness and death worldwide, but also provide the start for many other chronic and deadly maladies from shingles to certain cancers. Recent viral research articles utilizing high content include tracking the process of reactivation of Kaposi’s sarcoma-associated herpesvirus6 and screening a library for antiviral activity against JC polyomavirus (causing progressive multifocal leukoencephalopathy [PML]).7 Remarkably, this small screen (2000 compounds) identified mefloquine, an antimalarial agent, as a potentially effective PML therapy.
Neurodegenerative diseases such as Alzheimer’s, Parkinson’s, Huntington’s, amyotrophic lateral sclerosis, and brain cancer are global health issues, robbing the aging population of quality of life and taking an enormous financial and emotional toll on families. An understanding of the basic mechanisms involved in neuronal development, function, and death is paramount to developing new treatments, and high content is again playing a role. The RIKEN Brain Sciences Institute in Saitama, Japan, used the ArrayScan VTI (Thermo Scientific Cellomics™) to study Rho kinase-enhanced degradation of toxic aggregates resulting from mutant Huntington genes.8 More than 10,000 individual cells per well were automatically counted and quantified for the presence of inclusions in the study.
It is the ability of HCS to measure morphology of developing neurons that has helped to revolutionize neuroscience therapeutic programs at pharmaceutical, biotechnology, and academic institutes. Indeed, measurement of the outgrowth of neurons was a landmark development in HCS and one that has spurred the growth of the technology.9,10
The ability to address phenotypes can also be used to build novel assays for in vitro toxicity assessment where subtle, multiparameter cellular profiles can provide a wealth of information on cell health. High content is currently utilized for a wide variety of applications, including identifying drug-induced liver injury (DILI);11 analysis of the potential toxicity of emerging materials, such as nanoparticles;12 and understanding the mechanisms of action of toxic components in consumer products, such as cigarettes.13
High content fosters collaboration
Eli Lilly and Company (Indianapolis, IN) recently announced the desire to increase collaboration with the biotechnology industry by exploiting the unique properties of high-content methodology. According to the press release, “the initiative, called the Lilly Phenotypic Drug Discovery Initiative or PD2 (pronounced PD-squared), uses Lilly-developed disease-state assays and a secure web portal to evaluate the therapeutic potential of compounds synthesized in university and biotechnology laboratories. Findings from this initiative could ultimately form the basis for collaboration or licensing agreements between Lilly and external institutions.” The high-content assays have been developed around four disease areas (Alzheimer’s, cancer, diabetes, and osteoporosis) and provide a way to better profile a compound’s pharmacology. External consultant for the project, Dr. Peter Wipf, Ph.D., from the University of Pittsburgh, has written for several publications about the role of high-content technology in his research.14,15
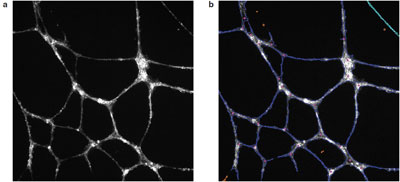
Figure 1 - Angiogenic tube formation in Huvec cells: a) raw image, and b) algorithmic overlays from Cellomics BioApplication image analysis software. F-actin and tubulin staining were done using the Cellomics Cytoskeletal Rearrangment Kit.
In the oncology segment of the initiative, several examples of high content use can be found, starting with angiogenesis. Neovascularization of solid tumors is a critical step in the development of many cancers, and the ability to block this can starve the tumor of nutrients, giving surgical, chemotherapy, or radiation intervention a greater chance of success. The traditional approaches to measuring this phenomenon are either very indirect or very manual. High content, however, can image and quantify these tubes (Figure 1) in an automated and high-throughput fashion, providing information on tube size and shape, connectedness, as well as measurement of cell signaling occurring within tube itself. Lilly scientists recently assessed 47 different morphological endpoints for tube formation, identifying six parameters that were then cross-correlated with microarray studies to identify the involvement of various oncogenes in angiogenesis.16
Another study dissected the eukaryotic cell cycle into individual cellular subpopulations using high-content imaging (HCI) in conjunction with unsupervised K-means clustering. Distinct phenotypic fingerprints for each major cell cycle and mitotic compartment were used to screen a library of 310 commercially available chemotherapeutic agents.17
A third study looking into eIF4E activation levels in human prostate cancers used high content for simultaneous multiplexed apoptosis (nuclear morphology, caspase-3, and terminal deoxynucleotidyl transferase dUTP nick end labeling [TUNEL]), cell cycle, and eIF4E expression levels in cultured prostate cancer cells.18 Low et al. also published a review of phenotypic screening focusing on various approaches for hit prioritization, including compound profiling, studying small-molecule effects with RNAi, pathway mapping, and chemical clustering.19
Summary
As high content celebrates its 10th anniversary in 2009, it is clear that the ability to understand the effects of drugs on cells using such techniques is having a tangible impact on the biotechnology industry. Productivity through innovation and collaboration will be the cornerstone of future success, and the ThermoScientific Cellomics high-content platform will continue to provide opportunities for both. By bringing scientists together and giving them more efficient tools, we have the best chance of turning more discoveries into new therapies.
References
- Zhang, J. The impact of the financial crisis on the pharma and biotech industries. Life Science Leader 2009, 1(5), 42–4.
- Chan, E.; Longatti, A.; McKnight, N.; Tooze, S. Kinase-inactivated ULK proteins inhibit autophagy via their conserved c-terminal domains using an atg13-independent mechanism. Mol. Cell. Biol. 2009, 29(1), 157–71.
- Mutka, S.; Yang, W.; Dong, S.; Ward, S.; Craig, D.; Timmermans, P.; Murli, S. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009, 69(2), 510–17.
- Soncini, C.; Carpinelli, P.; Gianellini, L.; Fancelli, D.; Vianello, P.; Rusconi, L.; Storici, P.; Zugnoni, P.; Pesenti, E.; Croci, V.; Ceruti, R.; Giorgini, M.; Cappella, P.; Ballinari, D.; Sola, F.; Varasi, M.; Bravo, R.; Moll, J. PHA-680632, a novel aurora kinase inhibitor with potent antitumoral activity. Clin. Cancer Res. 2006, 12, 4080–9.
- O’Brien, C.; Cavet, G.; Pandita, A.; Hu, X.; Haydu, L.; Mohan, S.; Toy, K.; Sanchez Rivers, C.; Modrusan, Z.; Amler, L.; Lackner, M. Functional genomics identifies ABCC3 as a mediator of taxane resistance in HER2-amplified breast cancer. Cancer Res. 2008, 68(13), 5380–9.
- Cheng, F.; Weidner-Glunde, M.; Varjosalo, M.; Rainio, E.; Lehtonen, A.; Schulz, T.; Koskinen, P.; Taipale, J.; Ojala, P. KSHV reactivation from latency requires pim-1 and pim-3 kinases to inactivate the latency-associated nuclear antigen LANA. PLoS Pathogens 2009, 5(3), e1000324.
- Brickelmaier, M.; Lugovskoy, A.; Kartikeyan, R.; Reviriego-Mendoza, M.; Allaire, N.; Simon, K.; Frisque, R.; Gorelik, L. Identification and characterization of mefloquine efficacy against JC virus in vitro. Antimicrob. Agents Chemother. 2009, 53, 1840–9.
- Bauer, P.; Wong, H.; Oyama, F.; Goswami, A.; Okuno, M.; Kino, Y.; Miyazaki, H.; Nukina, N. Inhibition of rho-kinases enhances the degradation of mutant huntington. J. Biol. Chem. 2009, 284, 13,153–64.
- Williams, G.; Wood, A.; Williams, E.; Gao, Y.; Mercado, M.; Katz, A.; Joseph-McCarthy, D.; Bates, B.; Ling, H.; Aulabaugh, A.; Zaccardi, J.; Xie, Y.; Pangalos, M.; Walsh, F.; Doherty, P. Ganglioside inhibition of neurite outgrowth requires nogo receptor function: identification of interaction sites and development of novel antagonists. J. Biol. Chem. 2008, 283, 16,641–52.
- Radio, N.; Breier, J.; Shafer, T.; Mundy, W. Assessment of chemical effects on neurite outgrowth in PC12 cells using high content screening. Toxicol. Sci. 2008, 105, 106–18.
- Xu, J.; Henstock, P.; Dunn, M.; Smith, A.; Chabot, J.; de Graaf, D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008, 105, 97–105.
- Byrne, F.; Prina-Mello, A.; Whelan, A.; Mohamed, B.; Davies, A.; Gun’ko, Y.; Coey, J.; Volkov, Y. High-content analysis of the biocompatibility of nickel nanowires. J. Magn. Magn. Mater. 2009; doi: 10.1016/j.jmmm.2009.02.035.
- Carter, C.; Hamm, J. Multiplexed quantitative high content screening reveals that cigarette smoke condensate induces changes in cell structure and function through alterations in cell signaling pathways in human bronchial cells. Toxicology 2009, 261, 89–102.
- Vogt, A.; McDonald, P.; Tamewitz, A.; Sikorski, R.; Wipf, P.; Skoko, J.; Lazo, J. A cell-active inhibitor of mitogen-activated protein kinase phosphatases restores paclitaxel-induced apoptosis in dexamethasone-protected cancer cells. Mol. Cancer Ther. 2008, 7, 330–40.
- Brisson Tierno, M.; Kitchens, C.; Petrik, B.; Graham, T.; Wipf, P.; Xu, F.; Saunders, W.; Raccor, B.; Balachandran, R.; Day, B.; Stout, J.; Walczak, C.; Ducruet, A.; Reese, C.; Lazo, J. Microtubule binding and disruption and induction of premature senescence by disorazole C1. J. Pharmacol. Exp. Ther. 2009, 328, 715–22.
- Chen, Y.; Wei, T.; Yan, L.; Lawrence, F.; Qian, H.R.; Burkholder, T.; Starling, J.; Yingling, J.; Shou, J. Developing and applying a gene functional association network for anti-angiogenesis kinase inhibitor activity assessment in an angiogenesis co-culture model. BMC Genomics 2008; 9:264; doi:10.1186/1471-2164-9-264.
- Low, J.; Huang, S.; Blosser, W.; Dowless, M.; Burch, J.; Neubauer, B.; Stancato, L. High-content imaging characterization of cell cycle therapeutics through in vitro and in vivo subpopulation analysis. Mol. Cancer. Ther. 2008, 7, 2455–63.
- Graff, J.; Konicek, B.; Lynch, R.; Dumstorf, C.; Dowless, M.; McNulty, A.; Parsons, S.; Brail, L.; Colligan, B.; Koop, J.; Hurst, B.; Deddens, J.; Neubauer, B.; Stancato, L.; Carter, H.; Douglass, L.; Carter, J. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009, 69(9), 3866–73.
- Low, J.; Stancato, L.; Lee, J.; Sutherland, J. Prioritizing hits from phenotypic highcontent screens. Current Opin. Drug Disc. Dev. 2008, 11(3), 338–45.
Mr. Zock is Market Segment Manager, Cellular Imaging and Analysis, Thermo Fisher Scientific, 100 Technology Dr., Pittsburgh, PA 15219, U.S.A.; tel.: 412-770-2500; fax: 412-770-2450; e-mail: [email protected].