Over the past 15 years, label-free, real-time biomolecular interaction analysis has established itself across a broad spectrum of disciplines from drug discovery to basic life science R&D.1,2 The field has also seen an increase in the number of instrumentation providers bringing this powerful technique to almost all segments of the scientific community. There is a strong trend toward increasing throughput with the incorporation of higher numbers of sensing channels and parallel sample processing.3,4 This increases the complexity of these systems and adds significantly to the cost of the instrumentation. However, automation has enabled extremely reproducible sample handling, and when combined with double referencing of interaction data sets, a significant improvement in almost all performance metrics is obtained. These elements are integral to the performance of the SensíQ Pioneer® high-performance surface plasmon resonance (SPR)-based biosensing system (Icx Nomadics, Oklahoma City, OK).
Microtiter plate handling and autosampler-based sample processing (e.g., dilution preparation, mixing, transfers) are now considered standard elements of any high-quality system. There is, however, a balance between throughput, data quality, and cost that must be considered. Adding parallel sensing channels is not the only way to improve throughput. In fact, SensíQ Pioneer utilizes a highly reproducible single-probe autosampler/injector system to form a continuous gradient in analyte concentration as the analyte is delivered along a microfluidic path to the sensing channels. This continuous analyte gradient injection encompasses a wide concentration range and is intended to replace a complete conventional analyte dilution set. Typically an analyte dilution set covers 2–3 orders of magnitude and will require 6–10 vial positions in the sample rack. By eliminating the need for such dilutions, it is possible to kinetically screen 192 different analyte samples in a single assay run without performing any provisional experiments to find the optimal concentration range for each analyte. The associated reduction in experimental complexity and assay time enables this serial sampling system to rival more complex instruments with parallel sampling capabilities while providing higher data quality. In many cases, the addition of parallel sensing channels (>20) can only be accomplished at the expense of sensitivity and may negate the benefit afforded by increased parallelism.
Sensitivity is the most critical performance metric when considering screening and kinetic characterization of low-molecular-weight chemical entities. SensíQ Pioneer employs an SPR detector that is sensitive to less than 1 × 10–7 refractive index units and can monitor fast binding/dissociation events of very small (<100 Da) or large analytes (>1-μm particles) over a very wide affinity range (pM to mM).
What are the key performance features that can be used when comparing instruments? The critical assessment of competing instruments can be a difficult prospect for even experienced scientists. The many systems currently available have individual strengths and weaknesses that must be weighed on a cost/performance basis and consideration of the individual demands of the application of interest. However, there are several performance features that can be used to reliably rank the quality of any system. A scientist experienced in the analysis of binding response curves will usually be able to discern high-quality data from poor data in a single glance, but this is not trivial for the new user. This article elaborates on the more important performance features that indicate data quality by using data sets recorded on the SensíQ Pioneer biosensing system.
General data quality
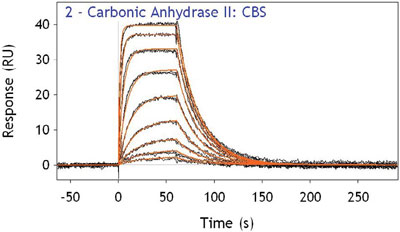
Figure 1 - Overlaid binding response curves for interaction of CBS with immobilized carbonic anhydrase II. Kinetic constants ka 2.3 × 10–4M-1 sec–1 and kd 0.04 sec–1 were obtained from the kinetic model fit.
The interaction of the inhibitor carboxybenzenesulfonamide (CBS) with the immobilized enzyme carbonic anhydrase II is shown in Figure 1. Binding interaction curves for nine different concentrations of CBS, in duplicate, are shown. The duplicates are completely superimposable, indicating extremely good reproducibility. CBS is a low-molecular-weight analyte, yet the data show a high signal-to-noise ratio (400:1). This performance is for the most part dependent on the detector limit of detection and the capacity/biocompatibility of the biosensor chemistry. A stable baseline free from undulations before the sample injection indicates good-quality reference curve subtraction and low thermal drift. The plateau in the response at each concentration indicates that a steady-state condition exists for the complex, but also suggests that overcrowding (i.e., hindered accessibility of analyte to immobilized enzyme) is likely to be absent. However, the greatest indicator of data quality is the almost perfect agreement between the kinetic model fit (red curves) and the data (black curves). This interaction is known to obey the simple 1:1 interaction model; thus a global fit of the kinetic parameters (ka, kd, Rmax) should fit closely unless there are artifacts that prevent this. Mass transport limitation, analyte–ligand overcrowding, rebinding effects, poor reference curve subtraction, analyte depletion, analyte dispersion, and large fluidic dead volumes are some of the more common artifacts.2 The occurrence of such artifacts may in some cases be linked with incorrect experimental setup, but in other cases may be inherent to the instrument.
Monitoring fast interaction events
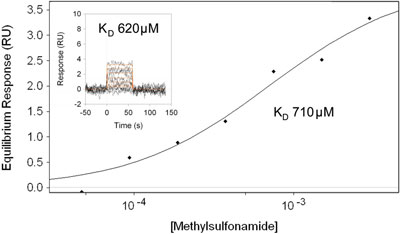
Figure 2 - Steady-state model fit for analysis of methylsulfonamide with immobilized carbonic anhydrase II. The inset is the kinetic model fit for the same data set.
A steady-state model fit for the interaction of the inhibitor methylsulfonamide (molecular weight 95 Da) with the immobilized enzyme carbonic anhydrase II is shown in Figure 2. The inset shows the kinetic curves with the associated affinity constant from the kinetic analysis. In this case, the dissociation rate constant (kd) for the interaction was extremely fast (i.e., 2 sec–1). The accurate resolution of this rapid event is enabled by an advanced fluidic design that causes the transition from buffer to analyte to occur within 1 sec during a typical injection. Normally this is not possible with conventional flow injection systems, and this limits such systems to a narrower kinetic measuring range. The relatively good agreement between the steady-state model and the kinetic model fit is a strong indicator of a high-quality data set.
Mass transport limitation
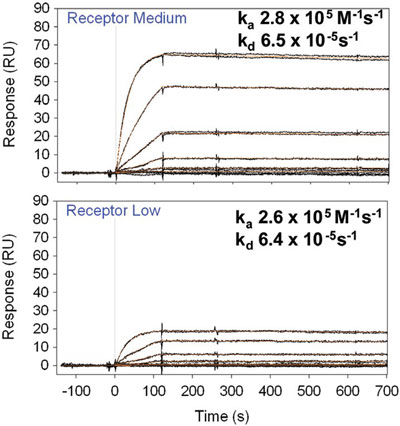
Figure 3 - Overlaid binding response curve for the interaction of scFv with affinity-captured receptor on two working surfaces with different quantities of immobilized receptor.
Nanoliter-scale flow cells are required to support a high mass transport flux of analyte from the bulk liquid to the interaction surface. In contrast, large-volume flow cells give rise to transport limited response curves that increase the error in the fitted kinetic constants. Figure 3 shows two high-affinity data sets from SensíQ Pioneer for the interaction of an immobilized receptor (affinity captured onto a Protein G-coated surface) with an scFv (molecular weight of 27 kDa). Each plot shows binding response curves for six serial tripling dilutions, in duplicate, over two sensing channels that have been coated with different quantities of receptor. The global fit of the 1:1 interaction model is almost perfectly superimposable onto both data sets and returns very similar kinetic values despite the large difference in the binding capacity for each sensing channel. It can be concluded that transport limitation is absent since the presence of mass transport limitation would have been revealed as a systemic deviation from the model that would be more severe for the highercapacity surface.
Conclusion
SensíQ Pioneer is an automated, three-channel, flow-injection-based SPR system that employs serial sampling of analyte from vials and standard microtiter plates. The combination of advanced microfluidics, tailored surface chemistry, high detector sensitivity, temperature control, a precision autosampler, and method programming results in high data quality. In particular, affinity binding pairs that are difficult or impossible to monitor by other means can often be successfully detected and analyzed. In addition, the instrument throughput is sufficient to allow broad applicability in the field.
References
- Boozer, C.; Kim, G.; Cong, S.; Guan, H.W.; Londergan, T. Looking towards label-free biomolecular interaction analysis in a high-throughput format: a review of new surface plasmon resonance technologies. Curr. Opin. Biotechnol. 2006, 17, 400–5.
- Rich, R.; Myszka, D.G. Survey of the year 2007 commercial optical biosensor literature. J. Mol. Recognit. 2008, 21, 355–400.
- Choi, C.J.; Cunningham, B.T. A 96-well microplate incorporating a replica molded microfluidic network integrated with photonic crystal biosensors for high throughput kinetic biomolecular interaction analysis. Lab Chip 2007, 7, 550–6.
- VanWiggeren, G.D.; Bynum, M.A.; Ertel, J.P.; Jefferson, S.; Robotti, K.M.; Thrush, E.P.; Baney, D.M.; Killeen, K.P. A novel optical method providing for high-sensitivity and high-throughput biomolecular interaction analysis. Sens. Actuators B. 2007, 127, 341–9.
Mr. Havard, Mr. Gillock, and Mr. Martin are Research Scientists, and Dr. Quinn is Chief Scientist of Bioinstrumentation, Icx Nomadics, 800 Research Pkwy., Ste. 100, Oklahoma City, OK 73104, U.S.A.; tel.: 405-239-8600; fax: 405-235-8608; e-mail: [email protected].