Quantification of the kinetic rates of protein association, dissociation, and aggregation is an essential factor in understanding and ultimately manipulating these phenomena. A variety of measurement techniques are commonly applied, yet a given system may or may not be amenable to monitoring via any of these detection methods. Standard enzymatic spectrafluorescence or radiometric assays require labeling the molecules of interest, bringing into question the effect of the modification on the reaction rates. Selection of an appropriate substrate also holds the potential for confounding the measurement. Biophysical assays such as surface plasmon resonance (SPR) require receptor immobilization on a surface, again with the potential for modifying the reaction relative to a true free-solution measurement. More generally, these techniques provide only an indirect measure of the actual physical phenomenon of interest. For example, while fluorescence may be well-correlated to complex formation, the fluorescence signal itself is not a direct measure of an inherent property of the complex.
CG-MALS and TD-MALS
Multiangle, static light scattering (MALS) is a widespread technology for characterizing macromolecules in solution, providing an absolute measure of molar mass and size. Typically, MALS detectors are coupled to a separation technique such as size exclusion chromatography (SEC-MALS) or field flow fractionation (FFF-MALS) for determining the distribution of molecules in solution by molar mass, without reference to standards. A more recently developed technique is automated composition gradient, or CG-MALS, wherein equilibrium macromolecular interactions are quantified by measuring the light scattering signal over a series of compositions. As described previously,1 CG-MALS can determine equilibrium association constants and stoichiometries of reversibly associating protein complexes, both homo- and heteroassociations. CG-MALS measurements do not require fluorescent or radioactive tagging, additional substrates, or immobilization, and thus access free-solution properties of unmodified samples in almost any solution. Some of the more promising applications of CG-MALS in the pharmaceutical industry include antibody/antigen binding, small-molecule inhibition, liposome uptake of proteins, and antibody formulations. Fundamental studies of chaperon proteins, enzyme/inhibitor interactions, binding of protein–surfactant or DNA–protein systems, and so forth will also benefit from this technique.
An essential advantage of static light scattering over some other methods is its ability to determine the molar masses of the complexes and constituents directly from the measured signals. Analysis of the CG-MALS data yields the stoichiometry of complexes formed, as well as the association constants.
A natural extension of CG-MALS is time-dependent static light scattering, or TD-MALS, wherein the intensity of light scattered by a solution is monitored over time, after a rapid step of preparation and delivery to the detectors. While not new, the application of static light scattering to protein–protein interaction kinetics has been fairly limited. Some of the systems studied via stopped flow static light scattering include dimer–tetramer equilibration of human hemoglobin upon rapid pH changes,2 ribosome dissociation and subunit association,3 viral matrix protein binding to host nucleocapsids,4 DNA/poly-L-lysine polyplex formation,5 and fibrinogen formation.6 The potential applications of TD-MALS include not only fundamental molecular biology but also investigation of reversible and irreversible aggregation of therapeutic proteins in formulation buffers.
Automation
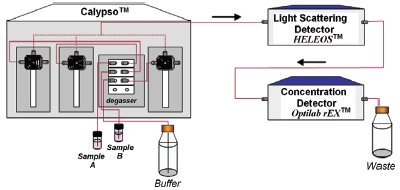
Figure 1 - Schematic TD-MALS setup, including the Calypso triple-syringe pump accessory, DAWN-HELEOS static light scattering detector, and Optilab rEX differential refractometer.
In the TD-MALS method, a stopped-flow measurement is performed over a series of conditions. A TD-MALS procedure consists of preparing a series of solutions, delivering each in turn to both MALS and concentration detectors over a time scale that is short in comparison to the relaxation time, acquiring the data, and determining the reaction rate under each condition. The total time span of the experiment may be as short as a few minutes or as long as many hours. Depending on the number of desired conditions and the required data acquisition time, the experiment may become tedious, time-consuming, and prone to operator error. The Calypso™ system (Wyatt Technology Corp., Santa Barbara, CA) overcomes the difficulties of manual TD-MALS by automating the entire procedure, integrating sample preparation and delivery with data acquisition and analysis in one comprehensive package. The system works in conjunction with a Wyatt MALS detector, such as the DAWN®- HELEOS™ or miniDAWN®-TREOS™, and an optional on-line concentration detector such as the Wyatt Optilab® rEX™ differential refractometer or a third-party UV absorption detector, to provide a complete TD-MALS setup (see Figure 1).
Inhibition kinetics
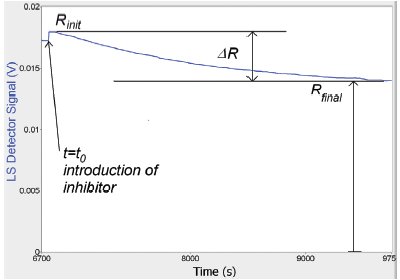
Figure 2 - Time-dependent behavior of light scattering signal from monomer–dimer–inhibitor solution. Upon introduction of the inhibitor molecule, the signal decreases exponentially from Rinitto Rfinalwith time constant τ.
This paper demonstrates the use of an automated TD-MALS system for characterizing the kinetics of dissociation of a protein complex. Alpha-chymotrypsin is known to reversibly self-associate at low pH, forming a monomer–dimer equilibrium.7 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) is an irreversible serine protease inhibitor that binds to the chymotrypsin active site,8 which is also the binding site for self-association. Upon adding AEBSF to a chymotrypsin solution, the inhibitor will gradually diffuse into the dimers and bind to the active site, causing the dimers to break up into monomers. If the inhibitor molarity is much larger than that of the chymotrypsin, the drop in dimer concentration follows an exponential time dependence, until all of the chymotrypsin dimers have dissociated. The accompanying decrease in the weight-averaged molar mass of the chymotrypsin solution is reflected in the TD-MALS signal, as shown in Figure 2.
For a single species in solution under dilute (“ideal”) conditions, the light scattering signal R(t) is proportional to the product of molar mass M and weight concentration c:
where K is a function of system parameters such as wavelength and solvent refractive index, and dn/dc is the sample refractive increment. If more than one species is present, then the total light scattering is just the sum of contributions from each species. The small molar mass of AEBSF relative to chymotrypsin ensures that the light scattering signal directly reflects the monomer–dimer balance, regardless of a small fraction of the inhibitor (which is present in a large molar excess) binding to the chymotrypsin.