Molds can cause health problems such as infections and allergies, destroy crops, and contaminate food or pharmaceuticals. They cannot be avoided. Molds are essential players in the biological processes on earth, but the molds that are most problematic can be identified and quantified, and removed in a timely manner. Through the recent development and application of the technique of mold-specific quantitative polymerase chain reaction (MSQPCR), indoor molds can now be identified accurately and with such quantifying precision that predictive models are emerging that can help define abnormal mold conditions.
How MSQPCR has advanced the detection and quantification of molds
MSQPCR is a highly accurate and sensitive molecular technique for the detection of molds. The previous standby method of identifying molds present in environmental samples depends on culturing molds on defined media, knowing that only a handful of those molds actually present in a sample may take to the culture medium and grow in the laboratory. Methods long in use for quantifying molds rely on visually counting mold spores, a very slow, laborious, and often subjective process dependent on the refined skill of the individuals performing the tests.
MSQPCR obviates the limitations of these century-old methods of detecting and quantifying molds. MSQPCR is objective and specific because it is a detection system based on unique DNA sequences.
Preparation of samples
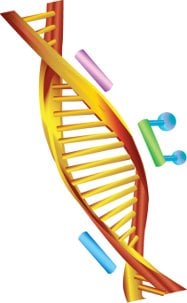
Figure 2 - Schematic of location on the DNA molecule for the MSQPCR-specific forward primer (pink), reverse primer (blue), and probe (green) with labeling dyes (shown as blue dots).

Figure 1 - Mini-Bead beater™ (Biospec Products, Bartlesville, OH) holding screw-cap tube (PGC Scientific, Gaithersburg, MD) used for DNA extraction of mold spores.
For assessing the prevalence of molds in indoor environments, generally either air or dust samples are collected. The DNA from such samples is obtained during the first step using a bead-beating extraction, which takes 1 min (Figure 1). For most air samples, the supernatant is ready for analysis following a brief centrifugation step. For dust samples, the supernatant is put through a purification step following centrifugation.
Unlike the original PCR process, quantitative PCR (QPCR, developed in 1997 by Applied Biosystems, Foster City, CA) adds a dual fluorescently labeled DNA probe to the original forward and reverse primers (Figure 2). The primers and probe are mixed with the proper enzymes and other reagents to accomplish the reaction. Primers and probes for a QPCR reaction are key components to the process and must be designed to be species-specific.
The MSQPCR process
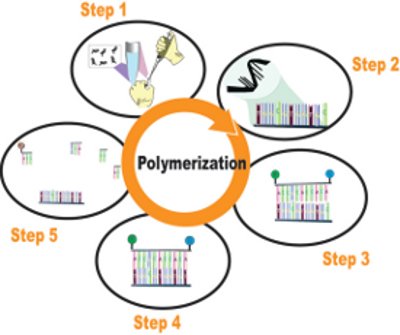
Figure 3 - Diagram of the MSQPCR amplification process. Step 1 is the mixing of reagents, step 2 is the creation of the single-stranded DNA, step 3 shows the case in which the probe does not match the target (note one base pair not a match), step 4 shows the case in which the probe matches the target, and step 5 demonstrates the separation of the dyes from the probe when a match is found.
As shown in Figure 3, a small aliquot of the sample DNA extract (supernatant) is added to an MSQPCR reaction mixture (step 1) containing the primers and probe for a specific mold. This process is repeated for each target mold species being assayed. Heating the DNA in the sequence detector thermocycler (step 2) causes the double helix to unwind, leaving single strands. The mold-specific primers and probe then interact with the sample DNA.
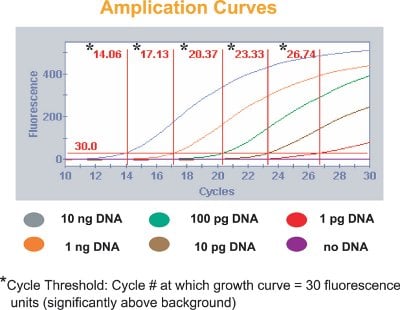
Figure 4 - Examples of the amplification curves generated by the sequence detector measuring the fluorescence from differing quantities of target DNA.
If there is no exactly matching sequence to the probe in the sample DNA (step 3), no signal is detected. When the mold-specific DNA fluorescent probe finds a match in the sample DNA (step 4), the fluorescent dye on the probe is released and measured by the sequence detector (step 5). The amount of fluorescence is directly proportional to the number of cells for each specific mold, thus making the process quantitative. In short, the number of mold spores is quantified, based on a standard calibration, from the number of cycles required to exceed a fluorescence threshold (Figure 4).
There are many manufacturers of sequence detectors; some of these instruments have been reviewed recently.1 Sequence detectors perform the heating and cooling of the reaction for the PCR part of the process, and they also monitor the fluorescence emitted from the prepared DNA probes. These instruments, which cost from about $30 to 80 K, vary in the number of reactions that can be run at one time, ranging from 1 to 384.
Application of MSQPCR in the study of homes
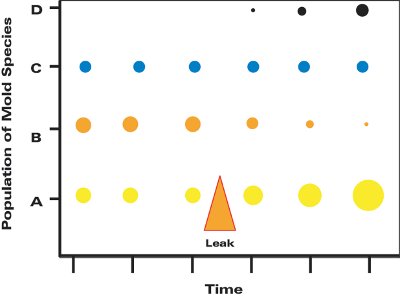
Figure 5 - Examples of possible changes in four mold populations after a water intrusion event, e.g., a leak (note orange triangle). Size of dots indicates relative population of each mold.
The application of MSQPCR that has been the authors’ focus relates to the understanding of the dynamics of indoor mold populations, especially in homes. There has been heightened attention drawn to the putative hazards of excessive mold growth within the structure of homes. When moisture conditions become abnormal in a home, some mold species increase in abundance, some decline, some remain the same, and even new mold populations may appear (Figure 5). The ecosystem of the indoor space, including the mold populations, changes. But since there is wide variation among homes, one cannot expect that all water-intruded homes or buildings will have exactly the same mold growth niches.
To understand the dynamics of changes in mold populations in a water-intrusion event or condition, the authors sought to develop a method that would allow mold population levels to be expressed in a generalized but predictive way. They needed a good approach to smooth out these variations in mold growth niches within different homes. The geometric mean (GM) of each mold concentration allowed that to be accomplished for 82 molds in home dust samples from visibly moldy homes compared to a control group.2,3 Therefore, in the authors’ application of MSQPCR to the study of mold conditions in homes, they use the GM of each mold population as the relevant method of grouping the molds.2,3
The U.S. EPA (Cincinnati, OH), in conjunction with Housing and Urban Development (HUD, Washington, D.C.), is currently undertaking a nationwide study of molds in homes across the U.S., scheduled to be completed in 2005. The MSQPCR technology is being used in this study. Obtaining the GM profiles of indicator species for a moldy home versus a “normal” home is the objective of this study. With this larger set of data, the authors will be testing more thoroughly and validating the predictive nature of the GM ratio profile approach.
Application of MSQPCR in hospital settings and in the food and pharmaceutical industries
There are many other applications for MSQPCR. In health-care settings, nosocomial fungal infections have a high fatality rate. Thus, the infection control specialist needs to be on the constant lookout for certain species of molds, especially of the genus Aspergillus. The sensitivity of MSQPCR is expediently useful in these settings. The surveillance for possible pathogenic fungi is particularly critical in hospitals during construction and renovation projects. The disruptive work of construction can stir up molds of all types, including those most notorious for causing infections. The real-time availability of data from MSQPCR can now assist hospital infection control staff in monitoring the presence of potentially pathogenic molds in the patients’ environment. 4,5 Molds are manageable, but we must monitor for them in a very timely fashion and act quickly to eliminate them.
In agriculture, molds are sources of mycotoxins that contaminate human and animal food. Molds also threaten and destroy crops. MSQPCR can be used to monitor plant pathogens and toxin-producing fungi in the field and in grain elevator storage of feed.6
Additional benefits of MSQPCR for environmental mold analysis
A very important advantage in using MSQPCR, in addition to the high specificity and quantification, is the rapidity with which the assays can be performed. Results can be obtained within approx. 3 hr or less. These three strengths of MSQPCR demonstrate its very significant capacity and usefulness compared to the weeks-long low specificity obtained using the century-old “standard” mold culture techniques. As MSQPCR gains momentum in the mold analysis field, we are on the threshold of significant advances in understanding and controlling damaging molds.
DNA probes and primers are available to provide assays for more than 100 of the most common problem mold species. The growth in the use of MSQPCR is greatly aided by the U.S. EPA, which is making this technology available through licensed companies in the U.S. and Europe.
References
- Pray LA. Consider the cycler. Scientist 2004; 18:34–37.
- Vesper SJ, Varma M, Wymer LJ, Dearborn DG, Sobolewski J, Haugland RA. Quantitative PCR analysis of fungi in dust from homes of infants who developed idiopathic pulmonary hemorrhaging. J Occup Environ Med 2004; 46:596–601.
- Meklin T, Haugland RA, Reponen T, et al. Quantitative PCR analysis of house dust can reveal abnormal mold conditions. J Environ Mon 2004; 6:615–20.
- Morrison J, Yang C, Lin K-T, Haugland RA, Neely AN, Vesper SJ. Monitoring Aspergillus species by quantitative PCR during construction of a multi-story hospital building. J Hosp Infec 2004; 57:85–7.
- Neely AN, Gallardo V, Barth E, Haugland RA, Warden G, Vesper SJ. Rapid monitoring by QPCR for pathogenic Aspergillus during carpet removal from a hospital. Infec Con Hosp Epi 2004; 25:350–52.
- Vesper S, Haugland RA. 21st century mold analysis in the food industry. Food Qual 2003; Nov/Dec:39–40.
Dr. Mary Jo Vesper is a freelance writer, MJVScript, Cincinnati, OH, U.S.A. Dr. Steve Vesper is Research Scientist, and Dr. Haugland is Molecular Microbiologist, U.S. Environmental Protection Agency, 26 W. M.L. King Dr., Cincinnati, OH 45268, U.S.A.; tel.: 513-569-7367; fax: 513-487-2512; email: [email protected]. Mr. Kahane is Principal, Forensic Analytical, Hayward, CA, U.S.A. The U.S. EPA’s Office of Research and Development partially funded and collaborated in the research described here. It has been subjected to the Agency’s peer review and has been approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use.