The genomic revolution of the last two decades has provided pharmaceutical companies with an almost overwhelming number of potential drug targets. The creation of potent new drugs addressing those targets is imminent. However, with most of the low-hanging fruit already picked, drug development is becoming an increasingly difficult endeavor. To gain FDA approval, new drugs must demonstrate safety and efficacy, especially when compared to existing drugs.
High-throughput methods such as microarrays for gene expression analysis and mass spectrometry for proteomic and metabolomic screening have had a huge impact on drug discovery and development over the past two decades. Consider:
- Drug targets are routinely identified using RNA microarrays
- Drug metabolism and pharmacokinetics are investigated using mass spectrometry measurements
- Diagnostic biomarkers are identified in the proteome using 2-D gels; mass spectrometry; and, more recently, protein arrays.
Many drugs that have been successfully developed target very specific pathways. This approach has worked well, but complicated diseases such as cancer often involve literally hundreds of genes. While diseases are traditionally classified by symptoms, many diseases with similar symptoms have very different origins at the genomic level and require different treatments. Transcript and protein biomarkers, especially when derived from readily accessible fluids such as blood and urine, are an ideal basis for developing diagnostic tests that can be used to predict disease progression.
However, they often fail to accurately predict and detect diseases that are rooted in genetic variations. Here, genetic variants such as single nucleotide polymorphisms (SNPs) and chromosomal aberrations can often be much more reliable predictors for specific diseases.
Given that many critical diseases originate in genomic variations, it is not surprising to see that efficacy of drugs is also highly dependent on genomic variations. This is actually an often-observed phenomenon in clinical trials: While a particular drug shows no significant improvements in the trial population as a whole, there are subgroups for which the new treatment shows vast improvements when compared to existing drugs. To pass the high thresholds generally required for new drugs, pharmaceutical companies strive to develop biomarker tests capable of identifying those responders prior to trial and treatment.
By the time a new drug reaches the market, high-throughput technologies have helped to generate vast quantities of biological profiling data. However, in many organizations, due to a combination of external and internal factors, data are not utilized to their fullest potential. Data have been acquired using a wide range of technologies and instrument vendors (e.g., gene expression microarrays from Affymetrix [Santa Clara, CA], Agilent [Santa Clara, CA], and Roche NimbleGen [Madison, WI]; mass spectrometry data from Bruker [Billerica, MA], LECO [St. Joseph, MI], Thermo Fisher Scientific [Waltham, MA], and Waters [Milford, MA]; and next-generation sequencing data from 454Life Sciences [Branford, CT], Illumina [San Diego, CA], and Life Technologies [Carlsbad, CA]), and require different preprocessing steps.
In vitro and in vivo experiments have used different animal models and patients, and to be comparable require mapping across technologies, biological entities (e.g., transcript, gene, protein), and organisms. Until recently, commercial software vendors did not provide data management and data analysis solutions capable of dealing with large and diverse datasets. In recent years, however, this problem has been addressed, and commercial software packages including Genedata Expressionist® (Genedata USA, Lexington, MA) support the management and integrated analysis of large and diverse biological data (Figure 1).

Figure 1 - Genedata Expressionist easily integrates data across technology platforms and instrument vendors.
In addition to the inherent data complexity, many organizations have strict divisions among research, development, and clinical trials. These divisions make it difficult to integrate data acquired in different stages of the drug design and development. Moreover, pharmaceutical companies increasingly rely on contract research organizations (CROs) to perform biological experiments and even clinical trials, with the preprocessing and analysis of the raw data also being outsourced to the CROs.
As seen in several large biomarker studies (e.g., Innomed-PredTox.org), it is often not a single marker but a combination of gene expression, copy-number variation, methylation, noncoding DNA, SNPs, and proteins that yields the high specificity required for predictive biomarkers. Thus, drug development organizations are beginning to realize the untapped potential of data from biological profiling experiments, and are looking to bring more data in-house for integrated analysis.
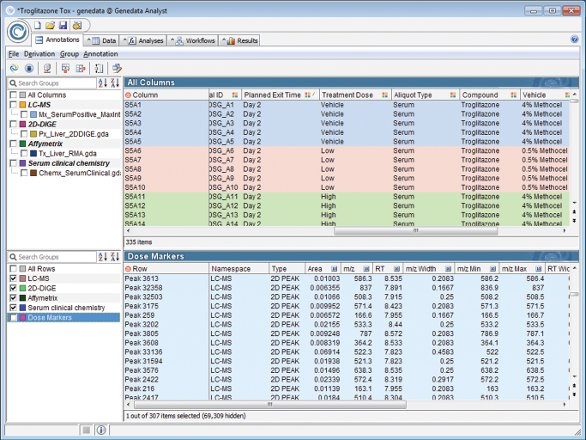
Figure 2 - Integrated analysis of InnoMed PredTox toxicogenomics data, combining different technologies and instrument vendors into a single dataset.
As a member of the InnoMed PredTox consortium, Genedata has developed data management and data analysis tools capable of integrating and analyzing data from a variety of different profiling technologies (Figure 2). Genedata scientists were part of the data analysis team that demonstrated that the combination of omics and conventional toxicology does indeed prove to be a useful tool for mechanistic investigations and the identification of putative biomarkers. Among others, the team identified novel biomarker sets and mechanisms of action for drug-induced liver toxicity within rats. Similar consortia are under way in Tucson, AZ, and Rockville, MD, at the Critical Path Institute (www.c-path.org ), and in Tokyo, Japan, at the ToxicoGenomics Project of the National Institute of Biomedical Innovation (http://toxico.nibio.go.jp/). Consortia results support the thesis of a true benefit in the integration of transcriptome, proteome, metabolome, and genome profiling data for, among others, toxicity prediction and patient stratification.
The rise of variant databases in drug development
The cost of gene expression, protein, and genomic profiling is rapidly dropping to levels that pale in comparison to the overall cost of clinical trials. With that in mind, the next logical step is to screen the transcriptome, proteome, and genome of all clinical trial participants using gene expression microarrays, mass spectrometry, and next-generation sequencing, respectively. The resulting data are combined with other profiling data and phenotypic information such as age, gender, and disease history into a single data repository called a variant database. Once sufficiently large variant databases are established, they will become an invaluable part of drug development and preclinical applications by enabling targeted meta-analysis correlating variants, phenotypes, and expression patterns. Researchers will be able to use variant databases to ask questions such as “Are certain genetic variations highly correlated with cardiovascular diseases?” or “Do certain genetic variations taken together with the presence of a particular protein in serum indicate that a patient will respond well to treatment?” Such a data repository will offer endless opportunities to drug development organizations, and increasingly companies will come to see them as an essential and valuable asset of their intellectual property.
Conclusion
Over the last two decades, molecular profiling technologies have found widespread adoption in drug discovery and development. Microarrays for gene expression analysis are used regularly for target identification and toxicity assessment; mass spectrometry has become a standard tool for studying drug metabolism and pharmacokinetics; and next-generation sequencing analysis is quickly gaining traction as a tool for variant analysis and patient stratification. Taken together, these high-throughput experiments yield vast amounts of data with data analysis akin to finding a biomarker needle in a huge haystack of data. Combining data from different profiling experiments across species and technologies will considerably enhance the level of detail and predictive power that these experiments yield.
The task of software providers is to develop the tools and databases capable of managing and analyzing these highly complex datasets combining expression profiling experiments, phenotypes, and genetic variants. Scientists need to be able to unlock the potential held by these experiments, and be ensured that data management and analysis will never be the bottleneck in the drug discovery and development process.
Dr. Hoefkens is Head of Genedata Expressionist®, Genedata USA, 750 Marrett Rd., One Cranberry Hill, Ste. 403, Lexington, MA 02421, U.S.A.; tel.: 781-357-4755; fax: 781-357-4770; e-mail: [email protected].