Microbiological testing is a large and expanding worldwide
market encompassing both research and industrial
sectors. In addition, the use of rapid molecular
methods for detection and identification, such as PCR
and sequencing, are increasingly becoming the standard
methods of choice. This adoption of rapid methods
means that enhanced formulations of reagents can allow
even untrained operators to set up PCR reactions in any
thermal cycler system, while still giving highly reproducible
results. Enhanced PCR reagent formulations such
as ReaX™ (Q Chip Ltd., Cardiff, Wales, U.K.) can also
offer experienced users easier PCR reaction setup and
high reproducibility by reducing the amount of pipetting
required. This is because all the reagents including
the primers, probes, Taq polymerase, buffer, and dNTPs
(2′-deoxynucleoside 5′-triphosphates) are encapsulated
into a single bead and only sample has to be added.
NCIMB Ltd. (Bucksburn, Aberdeen, U.K.), a specialist
microbiology company, maintains the largest industrial,
marine, and food culture collection in the U.K. and provides
services ranging from culture maintenance and
preservation to microbiological and chemical identification
and analysis. Sequencing of the 16S ribosomal RNA
(rRNA) gene is an important tool used by the company
for the rapid detection and identification of bacteria.
This investigation was designed to compare three methods
of PCR amplification of the 16S rDNA gene in order
to evaluate the performance and reliability of ReaX
Screwball Taq Mastermix beads relative to two other PCR methods. The work was carried out at NCIMB.
ReaX Screwball beads are an enhanced PCR reagent
formulation in which all the reagents required to
perform PCR, including the Taq polymerase, are
encapsulated in a single dose within the bead. Only
the addition of primers and genomic DNA template is required. The beads are made of soluble hydrogel
material, which instantly dissolves at 90 °C.
The beads are manufactured using a MicroPlant™
device (Q Chip Ltd.), a novel technology based on the
precise repeatable and scaleable manipulation of fluids in
microfluidic circuits. This technology enables the development
of beads from the hydrogel polymers that are
extremely uniform in size (less than 2% CV in bead size)
and contain a precise amount of encapsulated reagent.
Method
Three methods of PCR amplification were compared in
this investigation: ReaX Screwball Taq Mastermix beads,
MicroSeq 16S rDNA PCR kit (Applied Biosystems,
Foster City, CA), and conventional PCR. The reproducibility
of the ReaX PCR method was also evaluated in a
separate set of reactions using Pseudomona aeruginosa and
Escherichia coli as template DNA.
Genomic DNA from four organisms—P. aeruginosa
and E. coli (both common Gram-negative organisms),
Bacillus atrophaeus (a Gram-positive endospore-forming
bacterium), and Shewanella frigidimarina (a new and
unusual species)—was isolated and used as template
DNA. The genomic DNA extraction was carried out
using Ultra Prepman reagent (Applied Biosystems).
All PCR reactions were set up in duplicate according
to either conventional PCR protocol or the manufacturer’s
instructions. The primers pA and pE* were
used in both the ReaX Screwball and conventional
PCR reactions. A nontemplate negative control was
performed in parallel with each type of PCR reaction.
All PCR reactions were set up in duplicate according
to either conventional PCR protocol or the manufacturer’s
instructions. The primers pA and pE* were
used in both the ReaX Screwball and conventional
PCR reactions. A nontemplate negative control was
performed in parallel with each type of PCR reaction.
All PCR reactions were performed in a PTC-200 thermal
cycler (MJ Research, part of Bio-Rad Laboratories,
Hercules, CA) using an optimized program.
Three microliters of each PCR reaction
product was visualized on a 1% agarose gel.
For evaluation of ReaX reaction reproducibility,
PCR reactions were set up as above
but in triplicate using P. aeruginosa and E.
coli as template DNA. The PCR reaction
products were visualized on a 1% agarose gel.
Results
Figures 1 and 2 compare the PCR amplification
performance of ReaX Screwball
Taq Mastermix beads with MicroSeq
and the conventional PCR method in
the four organisms tested. Figures 3 and
4 demonstrate the reproducibility of the
ReaX Screwball Taq Mastermix beads for
the four organisms tested.
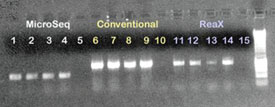
Figure 1 - 1% Agarose gel containing PCR products from PCR investigations
with P. aeruginosa and E. coli isolates using the MicroSeq method, conventional
PCR, and ReaX Screwball Mastermix beads along with dH2O negative controls.
Lanes 1, 2, 6, 7, 11, and 12: P. aeruginosa isolate; lanes 3, 4, 8, 9, 13, and
14: E. coli isolate; lanes 5, 10, and 15: negative control; lane 16: 1-kB ladder.
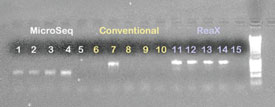
Figure 2 - 1% Agarose gel containing PCR products from PCR investigations
with B. atrophaeus and S. frigidimarina isolates using the MicroSeq method,
conventional PCR, and ReaX Screwball Mastermix beads along with dH2O negative
controls. Lanes 1, 2, 6, 7, 11, and 12: B. atrophaeus isolate; lanes 3, 4, 8,
9, 13, and 14: S. frigidimarina isolate; lanes 5, 10, and 15: negative control;
lane 16: 1-kB ladder.
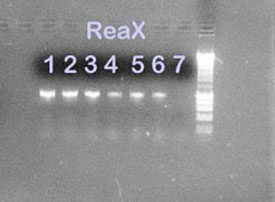
Figure 3 - 1% Agarose gel containing PCR amplification
products from P. aeruginosa and E. coli isolates using ReaX
Screwball Mastermix beads. Lanes 1, 2, and 3: P. aeruginosa
ReaX Screwball; lanes 4, 5, and 6: E. coli ReaX Screwball;
lane 7: ReaX Screwball negative control; lane 8: 1-kB ladder.
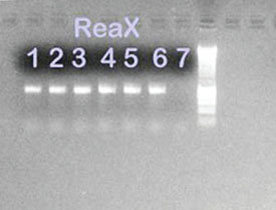
Figure 4 - 1% Agarose gel containing PCR amplification
products from B. atrophaeus and S. frigidimarina isolates
using ReaX Screwball Mastermix beads. Lanes 1, 2, and 3: B.
atrophaeus ReaX Screwball; lanes 4, 5, and 6: S. frigidimarina
ReaX Screwball; lane 7: ReaX Screwball negative control;
lane 8: 1-kB ladder.
Conclusion
For all organisms investigated (P.
aeruginosa, E. coli, B. atrophaeus, and S.
frigidimarina),
PCR reaction setup using
the ReaX Screwball Taq Mastermix beads
or the MicroSeq 16S rDNA PCR kit was
very user friendly, requiring no prior optimization,
and was quicker to perform compared
to the conventional PCR method.
Initially, the conventional PCR method
produced no bands but was successful when repeated and produced very intense bands. Repeating
this, however, was very time consuming. There were
no reaction failures using ReaX Screwball Taq Mastermix
beads or the MicroSeq 16S rDNA PCR kit.
ReaX Screwball Taq Mastermix beads reduced the
amount of manual pipetting required during PCR reaction
setup and therefore reduced the risk of repetitive
strain injury (RSI) over the long term. In the evaluation
of reproducibility, PCR bands generated using the beads
were identical for each triplicate and also for all four organisms.
These results indicated that the ReaX reagents give
highly reliable and reproducible PCRs for the purpose of
16S amplification and required no further optimization.
In conclusion, the ReaX Screwball Taq Mastermix beads
were found to be very user friendly and a much quicker
alternative to conventional PCR. They also demonstrated
to be very well optimized for the 16S assay, giving
reproducible results and minimizing pipetting error and
the opportunity for cross-contamination.
The authors are with NCIMB Ltd., Ferguson Bldg.,
Craibstone
Estate, Bucksburn, Aberdeen AB21 9YA, U.K.;
tel.: +44 0 1224 711100; fax: +44 0 1224 711299; e-mail:
[email protected].